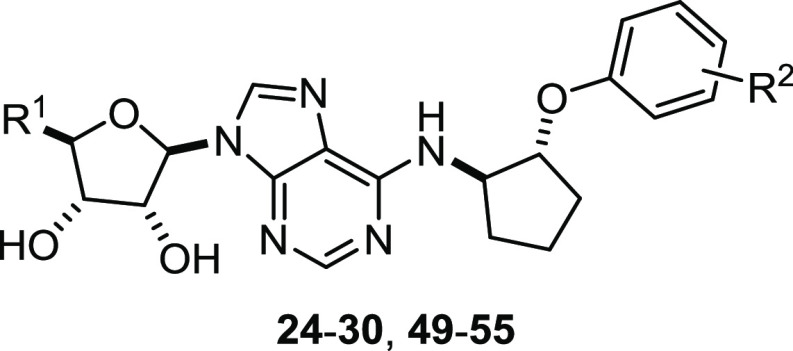
Table 5. Kinetics of Binding for Synthetic Adenosine and NECA Phenoxycyclopentyl Derivatives to the Orthosteric Binding Site at Human and Rat A1R.
| compd | R1 | R2 | hA1R |
rA1R |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| kon (k3) ×105 (M–1 min–1)a | koff (k4) (min–1)b | pKdc | RT (min)d | kon (k3) ×105 (M–1 min–1)a | koff (k4) (min–1)b | pKdc | RT (min)d | |||
| 24 | –CH2OH | H | 4.78 ± 0.93 | 0.048 ± 0.004 | 6.97 ± 0.06 | 21.56 ± 2.13 | 2.70 ± 0.34 | 0.053 ± 0.001 | 6.70 ± 0.06 | 18.91 ± 0.30 |
| 25 | –CH2OH | p-t-Bu | 1.48 ± 0.04 | 0.054 ± 0.005 | 6.44 ± 0.03 | 19.12 ± 2.01 | 4.84 ± 0.37 | 0.484 ± 0.173 | 6.17 ± 0.30 | 5.53 ± 3.31 |
| 26 | –CH2OH | m-OMe | 2.97 ± 0.27 | 0.037 ± 0.003 | 6.90 ± 0.01 | 27.61 ± 2.09 | 3.32 ± 0.52 | 0.085 ± 0.007 | 6.58 ± 0.03 | 12.06 ± 1.03 |
| 27 | –CH2OH | m-Br | 10.12 ± 1.29 | 0.038 ± 0.012 | 7.47 ± 0.09 | 33.13 ± 10.40 | 13.30 ± 1.21 | 0.050 ± 0.005 | 7.42 ± 0.03 | 20.40 ± 1.86 |
| 28 | –CH2OH | o-Cl | 11.61 ± 1.31 | 0.052 ± 0.004 | 7.34 ± 0.04 | 19.55 ± 1.48 | 17.92 ± 1.30 | 0.048 ± 0.005 | 7.58 ± 0.01 | 21.46 ± 2.08 |
| 29 | –CH2OH | m-Cl | 25.90 ± 2.14 | 0.040 ± 0.003 | 7.81 ± 0.02 | 25.37 ± 1.70 | 30.07 ± 4.02 | 0.071 ± 0.010 | 7.63 ± 0.12 | 15.20 ± 2.50 |
| 30 | –CH2OH | p-Cl | 1.01 ± 0.31 | 0.056 ± 0.009 | 6.22 ± 0.13 | 19.46 ± 3.34 | 2.51 ± 0.90 | 0.056 ± 0.001 | 6.58 ± 0.15 | 17.90 ± 0.25 |
| 49 | –CONHEt | H | 16.43 ± 2.81 | 0.028 ± 0.002 | 7.63 ± 0.01 | 32.82 ± 1.02 | 25.12 ± 2.08 | 0.037 ± 0.002 | 7.83 ± 0.05 | 27.61 ± 1.66 |
| 50 | –CONHEt | p-t-Bu | 1.53 ± 0.11 | 0.091 ± 0.010 | 6.23 ± 0.06 | 11.47 ± 1.48 | 6.29 ± 0.62 | 0.070 ± 0.007 | 6.95 ± 0.01 | 14.76 ± 1.64 |
| 51 | –CONHEt | m-OMe | 4.27 ± 0.03 | 0.027 ± 0.007 | 7.23 ± 0.11 | 42.68 ± 10.88 | 11.91 ± 1.33 | 0.030 ± 0.006 | 7.62 ± 0.11 | 38.05 ± 8.74 |
| 52 | –CONHEt | m-Br | 5.57 ± 2.56 | 0.075 ± 0.038 | 6.91 ± 0.04 | 27.87 ± 14.23 | 4.46 ± 1.28 | 0.056 ± 0.008 | 6.85 ± 0.13 | 19.17 ± 2.68 |
| 53 | –CONHEt | o-Cl | 17.27 ± 1.12 | 0.041 ± 0.010 | 7.66 ± 0.11 | 29.44 ± 6.91 | 37.59 ± 1.91 | 0.048 ± 0.003 | 7.90 ± 0.00 | 21.23 ± 1.22 |
| 54 | –CONHEt | m-Cl | 20.28 ± 1.11 | 0.043 ± 0.013 | 7.75 ± 0.16 | 34.70 ± 13.43 | 35.85 ± 3.95 | 0.062 ± 0.007 | 7.76 ± 0.10 | 16.78 ± 2.22 |
| 55 | –CONHEt | p-Cl | 3.75 ± 1.04 | 0.076 ± 0.024 | 6.71 ± 0.03 | 17.48 ± 4.79 | 11.62 ± 0.26 | 0.064 ± 0.003 | 7.26 ± 0.01 | 15.65 ± 0.78 |
kon (k3) for ligands as determined using NanoBRET binding assays using either human or rat Nluc-A1R expressing HEK 293 cells and determined through fitting with the ″kinetics of competitive binding″ model.35
koff (k4) for ligands determined as in footnote a.
Kinetic dissociation constant (pKd) for each ligand as determined from koff/kon.
Residence time of each ligand as determined by the reciprocal of the koff. All data are the mean ± SEM of at least three independent repeats conducted in duplicate.