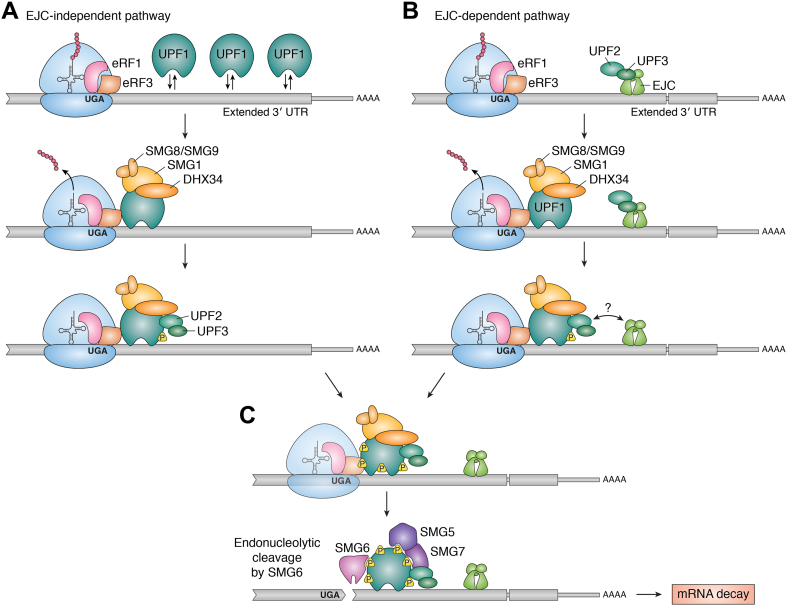
Figure 2.
NMD can occur in an EJC-dependent or EJC-independent manner. mRNA is shown in gray. The introduction of premature termination codon in coding sequence (thicker line) results in an “extended 3' UTR” that also includes the normal 3′ UTR (thinner line). The ribosome is shown in blue, and other important proteins are labeled. Recognition of NMD-inducing translation termination codon occurs through one of two general pathways: the EJC-independent (A) or EJC-dependent (B) pathway. A, the EJC-independent pathway begins when UPF1, the core NMD factor, is bound downstream of the terminating ribosome at abnormally high levels. After interaction between UPF1 and the termination complex, other NMD factors are recruited to assemble the “SURF” complex, which includes UPF1 phosphorylation factors SMG1 (the kinase) and its regulatory SMG8/9 heterodimer. This complex may include or subsequently recruit DHX34, an RNA helicase that may aid in interaction of UPF1 with its activators UPF2 and UPF3B (or its paralog UPF3A). Formation of the SURF complex leads to UPF1 phosphorylation (represented by yellow triangles labeled “P”) by SMG1. B, the EJC-dependent NMD requires the presence of an EJC downstream of the terminating ribosome that enhances NMD by recruiting UPF2 and UPF3B (or UPF3A) to mRNA. Following assembly of the SURF complex at the terminating ribosome and UPF1 interaction with its activators UPF2 and UPF3B (or UPF3A), which can be aided by DHX34, UPF1 gets phosphorylated by SMG1. C, after initial phosphorylation, UPF1 is phosphorylated more extensively. It is unknown how long the ribosome and release factors remain associated with the phosphorylated UPF1 (indicated by faded shapes). The extensive phosphorylation of UPF1 leads to the recruitment of SMG6 and SMG5/7 heterodimer, which initiate mRNA degradation via multiple mechanisms including SMG6-catalyzed mRNA endonucleolytic cleavage close to termination codons. EJC, exon junction complex; NMD, nonsense-mediated mRNA decay.