Fig. 1.
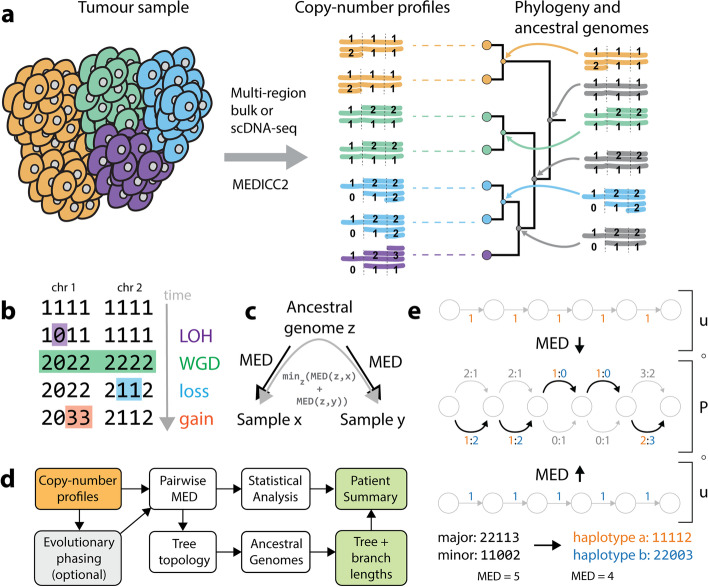
MEDICC2 algorithm. a MEDICC2 infers cancer phylogenies from SCNA data from single cells or bulk sequencing using a minimum-event distance (MED) and infers the ancestral genomes. It allows for backmutations, obeys biological constraints, and solves the phylogeny problem where ancestral genomes are not sampled. b Computing distances with WGD. Copy-number profiles are represented as vectors of positive integer copy numbers across chromosomes (here: two chromosomes with four segments each). To infer the correct MED, LOH events are considered first as lost segments cannot be re-gained by later events. WGD events span the full copy-number profile, whereas gain and loss events can affect an arbitrary number of segments within a chromosome. c Symmetric distance calculation. The MED from an ancestral state to a sample is asymmetric due to biological constraints. The final symmetric distance between two samples is computed as the sum of distances from an ancestral genome to both samples, while minimizing over all possible ancestors. d Schematic overview of the MEDICC2 workflow. Haplotype-specific copy-number profiles are either pre-phased or undergo evolutionary phasing (see e). Pairwise MEDs are computed between all genomes, followed by tree inference and ancestral reconstruction which determines the final branch lengths of the tree. Results are reported to the user as a patient summary and plot. e Evolutionary phasing. Copy-number profiles for both alleles are jointly encoded as an unweighted phasing FST P where both possible allele configurations are encoded at each position in the sequence. Evolutionary phasing then determines the optimal configuration (bold arrows) and extracts final haplotypes (orange and blue) by computing the MED between the phasing FST and two reference haplotypes. An example of major/minor copy number, phased copy number, and the MED from the diploid is shown at the bottom. Abbreviations: FST: Finite-state transducer, MED: Minimum-event distance, LOH: Loss-of-heterozygosity, WGD: Whole-genome doubling