Figure 2.
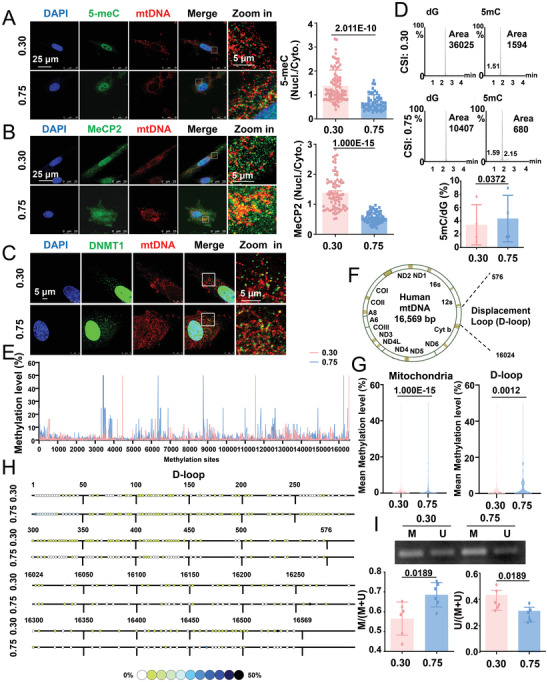
Geometric constraints regulate the methylation of mitochondrial DNA (mtDNA). A,B) Representative immunofluorescence staining and quantification of the nuclear/cytoplasmic ratio of 5‐methylcytosine (5‐meC) and MeCP2 in micropatterned SMCs. MtDNA was co‐stained with an anti‐mtDNA antibody. Each dot represents a single cell. C) Representative immunofluorescence staining of DNMT1 and mtDNA in micropatterned cells. D) Representative multiple reaction monitoring chromatograms of 5‐methyl‐deoxy‐cytidine (5‐mdC) and deoxyguanosine (dG) in the mtDNA of micropatterned cells. Quantitative statistical analysis of 5‐meC in micropatterned cells. Data were obtained from four biological repeats. E) Methylation patterns of the mtDNA of micropatterned cells. Schematic representation of the cytosine residue. Data were obtained from two duplicate samples. Each sample was obtained from four biological repeats. F) Schematic diagram of the human mitochondrial genome and D‐loop regions. G) The mean 5‐meC level of the entire mtDNA (left panel) and D‐loop (right panel) in micropatterned cells. H) Distribution of the D‐loop 5‐meC content in micropatterned cells. I) Representative agarose gel electrophoresis of the products of methylation‐specific PCR (MSP) for the D‐loop in bisulfite‐modified DNA isolated from the micropatterned cells. Semi‐quantification of the MSP results is shown in the lower panels. M: methylated, U: unmethylated. Immunofluorescent images were acquired using a confocal microscope. Significance was assessed using A,B) an unpaired t‐test, D,I) paired t‐test, or G) the Mann‐Whitney test. The error bars show ± SD. The exact P values between the indicated groups are presented.
