Abstract
Background
The yield of next-generation sequencing (NGS) added to a Sanger sequencing–based 16S ribosomal RNA (rRNA) gene polymerase chain reaction (PCR) assay was evaluated in clinical practice for diagnosis of bacterial infection.
Methods
PCR targeting the V1 to V3 regions of the 16S rRNA gene was performed, with amplified DNA submitted to Sanger sequencing and/or NGS (Illumina MiSeq) or reported as negative, depending on the cycle threshold value. A total of 2146 normally sterile tissues or body fluids were tested between August 2020 and March 2021. Clinical sensitivity was assessed in 579 patients from whom clinical data were available.
Results
Compared with Sanger sequencing alone (400 positive tests), positivity increased by 87% by adding NGS (347 added positive tests). Clinical sensitivity of the assay that incorporated NGS was 53%, which was higher than culture (42%, P < .001), with an impact on clinical decision-making in 14% of infected cases. Clinical sensitivity in the subgroup that received antibiotics at sampling was 41% for culture and 63% for the sequencing assay (P < .001).
Conclusions
Adding NGS to Sanger sequencing of the PCR-amplified 16S rRNA gene substantially improved test positivity. In the patient population studied, the assay was more sensitive than culture, especially in patients who had received antibiotic therapy.
Keywords: clinical metagenomics, 16S ribosomal RNA gene PCR, targeted metagenomics, tissue and body fluids
The addition of next-generation sequencing to 16S ribosomal RNA gene polymerase chain reaction/Sanger sequencing of normally sterile tissues and body fluids increased clinical sensitivity. In the study population, the described targeted metagenomic sequencing–based approach was more sensitive than culture, especially in patients who had received antimicrobial therapy prior to sampling.
Detection and identification of pathogens in infectious diseases are often based on conventional cultures followed by biochemical tests and/or, preferably, matrix-assisted laser desorption ionization time-of-flight mass spectrometry [1]. Cultures have poor sensitivity for fastidious organisms and/or with active or antecedent antimicrobial treatment [2]. This results in culture-negative infections that can be associated with overuse of unnecessarily broad-spectrum antibiotics, exposing patients to adverse effects of antibiotic therapy and selection of resistance or conversely untreated infections and/or delayed diagnoses [3–6]. The last can result in an unfavorable outcome, as shown for endophthalmitis, acute cholecystitis, and sepsis [7–9].
Molecular methods may circumvent these limitations by detecting microbial DNA directly in clinical samples. However, they are typically organism-specific. A broad-spectrum bacterial detection strategy is to target the 16S ribosomal RNA (rRNA) gene using polymerase chain reaction (PCR) followed by sequencing. Amplified product is then sequenced to identify the source bacterium. When Sanger sequencing is used with this approach, the overall positivity rate is similar to that of conventional cultures [10–12]. Sanger sequencing can generally only read one sequence at a time, restricting analyses to specimens that harbor single bacterial species and limiting sensitivity. In contrast, next-generation sequencing (NGS) can interrogate sequences of large numbers of bacteria at once, theoretically also increasing sensitivity [13]. This approach, an example of targeted metagenomic sequencing, has yielded promising results that suggest that addition of NGS can improve the positivity rate of 16S rRNA gene–based PCR/Sanger sequencing [14–16]. In contrast to shotgun metagenomics (sNGS), targeted metagenomics is not compromised of amplification of human sequences, is subject to facile bioinformatic analysis, and has a shorter turnaround time. However, large-scale experiences with this approach in routine microbiology laboratories have not yet been reported. To reduce labor and the cost of NGS, we developed and implemented in routine practice a targeted metagenomic sequencing–based approach (referred as tNGS) using 16S rRNA PCR followed by Sanger and/or NGS or direct reporting as negative, depending on the PCR cycle threshold (Ct) value. In this retrospective study, the yield of adding NGS to the 16S rRNA-based Sanger sequencing assay in clinical practice and the performance of tNGS compared with culture for infection diagnosis were assessed.
METHODS
Study Design
We conducted a retrospective study at a reference laboratory (Mayo Clinic Laboratories) to analyze samples received from 1 August 2020 (assay launch date) to 18 March 2021. Accepted samples were fresh or formalin-fixed paraffin-embedded (FFPE) tissues and body fluids collected from normally sterile sources.
Targeted Metagenomics Approach
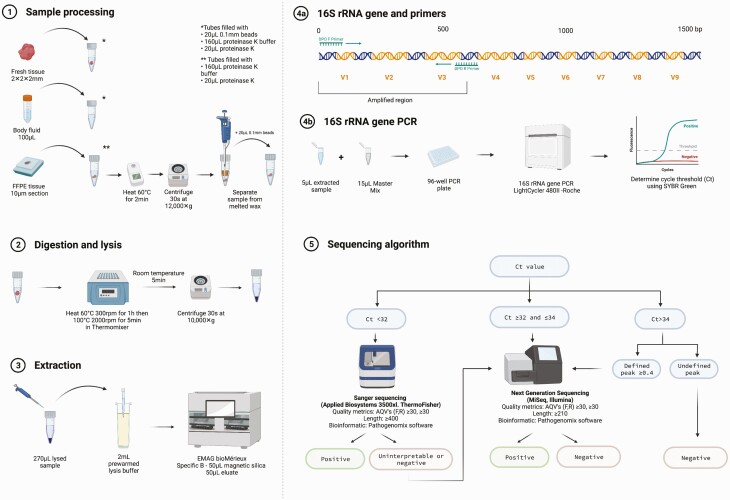
Specimens were processed, lysed, extracted, amplified, and sequenced as shown in Figure 1, along with negative, positive, and inhibition controls (Supplementary Methods). Amplification of the V1 to V3 region of the 16S rRNA gene was performed on a LightCycler 480II (Roche Diagnostics, Risch-Rotkreuz, Switzerland) using dual priming oligonucleotides, as previously described [17]. A Ct value was determined for each sample. Samples with Ct values <32 cycles were sent to Sanger sequencing. Those with Ct values ≥32 and ≤34 or <32 but yielding no identification, poor-quality results, or mixed chromatograms with Sanger sequencing were sent to NGS. Those with Ct values >34 cycles were considered negative and not further analyzed, except if a well-defined melting temperature (Tm) peak was observed, in which case they were sent to NGS.
Figure 1.
Laboratory workflow of the targeted metagenomics approach (tNGS). (1) A 2-mm3 piece of tissue or 100 µL of fluid was placed into a lysis tube with 20 µL of 0.1-mm silica/zirconium beads, 160 µL proteinase K buffer (PKB), and 20 µL of proteinase K. For FFPE tissues, a 10-µm section was put into a 1.5-mL centrifuge tube with 160 µL of PKB and 20 µL of proteinase K and heated to 60°C for 2 minutes to melt the wax. After centrifugation, liquid and tissue below the wax were transferred to a lysis tube with 20 µL of 0.1-mm beads. (2) Lysis tubes were incubated/spun, cooled for 5 minutes at room temperature, and then centrifuged. (3) Then, 2 mL of prewarmed (40°C) NUCLISENS easyMAG lysis buffer was added to the disposable NUCLISENS easyMAG cartridge with a maximum of 270 µL of the lysed sample and incubated at room temperature for 10 minutes followed by extraction loading. (4) Next, the 16S rRNA gene polymerase chain reaction targeted the V1–V3 region of the bacterial 16S rRNA gene using dual priming oligonucleotides. (5) Depending on the Ct value, samples were sent to Sanger or next-generation sequencing or reported negative. Abbreviations: AQV, average quality value; Ct, cycle threshold; FFPE, formalin-fixed paraffin-embedded; F,R, forward, reverse; PCR, polymerase chain reaction. Created with Biorender.com.
Bidirectional Sanger sequencing was performed on an Applied Biosystems 3500xl instrument (Thermo Fisher Scientific), and NGS was performed on an Illumina Miseq with a 500-cycle (2 × 250 paired-end read) v2 nano kit. Bioinformatic analysis was performed with RipSeq NGS software (Pathogenomix; Supplementary Methods). Doctoral-level clinical microbiologists (R. P. or N. W.) interpreted sequencing results. Organisms considered potential pathogens were reported. Organisms found in the negative control and/or as common assay background and/or across different samples in the same sequencing run were generally not reported.
Clinical Analysis
For clinical sensitivity analysis, samples from Mayo Clinic patients that underwent tNGS and culture were analyzed (Supplementary Figure 1). For both tNGS and cultures, if there was more than 1 sample from the same patient and same source sent to tNGS or culture, only 1 sample was counted, with the result considered positive if at least 1 of the replicates yielded a positive result (except for periprosthetic tissue cultures) and negative if all replicates from the same source were negative. For slow-growing organisms in periprosthetic tissue cultures, 2 positive cultures qualified as positive [18].
Patient and sample characteristics, including clinical and operative notes and laboratory and imaging results, were collected through electronic medical record review. Patients were classified as infected if a culture was positive (and not considered a contaminant) and/or if the treating medical team and the infectious diseases consult team considered the patient to be infected. Culture-negative cases with questionable infection status, observed off antibiotics with improvement were classified as uninfected.
Test impact on clinical decision-making was based on whether tNGS led to antimicrobial de-escalation or escalation and/or continuation or discontinuation of treatment, as recorded in the provider note.
Cultures
Conventional cultures included at least aerobic and anaerobic cultures and any other cultures (eg, fungal, mycobacterial) available. Periprosthetic tissue cultures were processed in aerobic and anaerobic blood culture bottles [19].
Statistics
Qualitative values were compared using the t test or Fisher exact test, as appropriate. Quantitative values were tested for normality using the Kolmogorov-Smirnov test and compared using Mann-Whitney or unpaired t tests, as appropriate. Clinical sensitivities, specificities, and negative and positive predictive values were calculated using 2 × 2 contingency tables, based on infection status. The 95% confidence intervals were calculated as exact binomial confidence intervals. To compare clinical performance, a McNemar test of paired proportions was performed. P values < 0.05 were considered statistically significant.
Ethics
The Mayo Clinic Institutional Review Board approved the study.
RESULTS
Samples
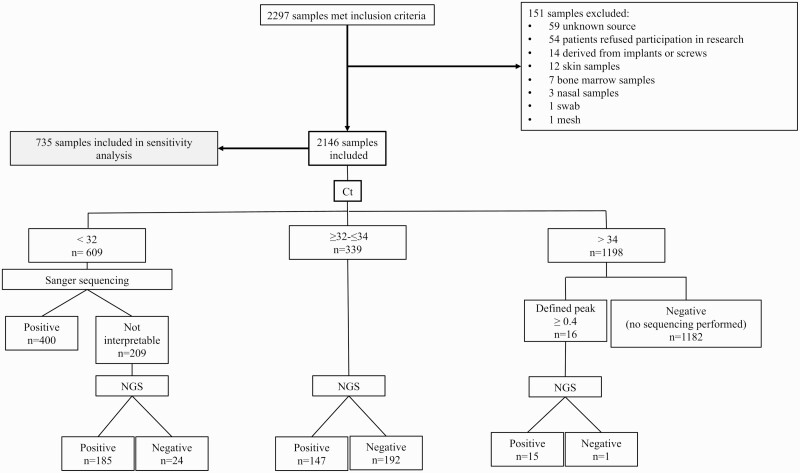
A total of 2297 samples met inclusion criteria, of which 151 were excluded. Among the 2146 included samples, 850 (40%) were fluids and 1296 (60%) were tissues (Figure 2). In the group with Ct values <32, 609 (28%) underwent Sanger sequencing, resulting in 400 (66%) positive and 209 (34%) uninterpretable results. The latter were sent for NGS, ultimately identifying a potential pathogen in 185 (89%) of the 209. The overall positivity rate in this subset was 96%.
Figure 2.
Sample flowchart. Skin and nasal biopsies were excluded as they are not normally sterile sources. Implants, screws, mesh, swabs, bone marrow, and samples of unknown source were excluded. Abbreviations: Ct, cycle threshold; NGS, next-generation sequencing.
A total of 339 (16%) samples had Ct values between 32 and 34, all of which underwent NGS, with 147 (43%) reported as positive and 192 (57%) negative. A total of 1198 (56%) samples yielded Ct values >34, of which only 16 underwent NGS, with positive and negative results reported in 15 (94%) and 1185 (99%), respectively, of samples in this group.
Additional Value of NGS
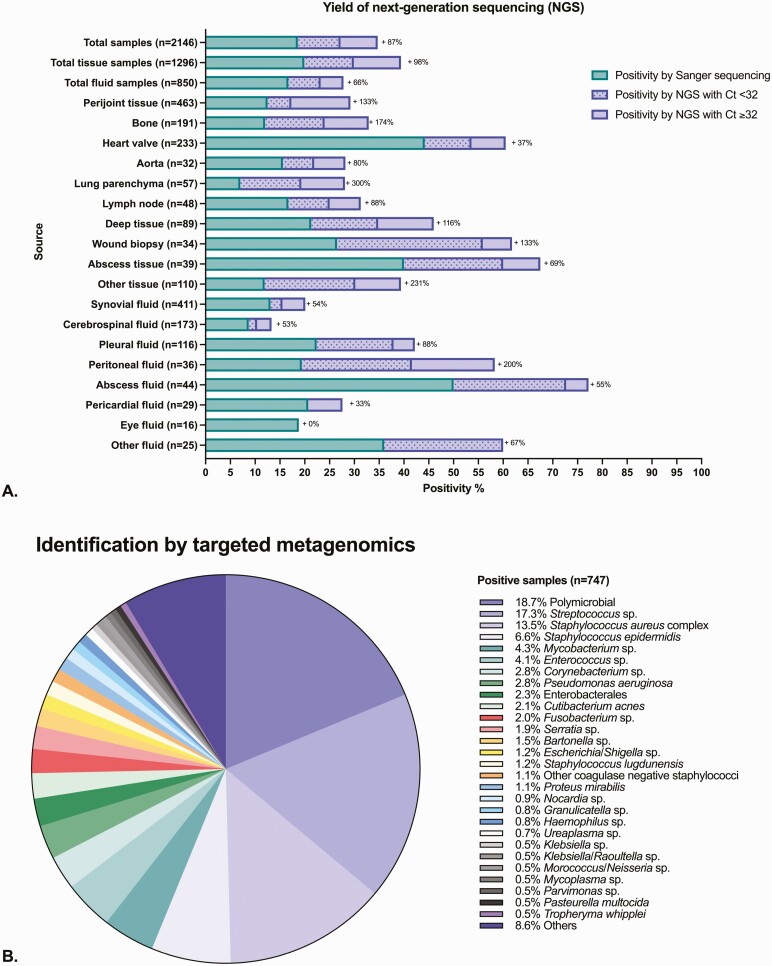
Overall, the sequencing-based assay was positive in 747 (35%) samples; in 400 (19%) by Sanger sequencing (all monomicrobial) and in 347 (16%) by NGS (140 [40%] polymicrobial and 207 [60%] monomicrobial).
NGS increased positivity by 87% compared with Sanger sequencing alone (98% for tissues and 66% for fluids; Figure 3A). Increased NGS yield compared with Sanger sequencing was highest for lung tissue (300%), peritoneal fluid (200%), other tissues (231%), bone (174%), and wound biopsies (133%) and was lowest for eye fluids (0%), pericardial fluids (33%), heart valves (37%), cerebrospinal fluid (53%), and synovial fluid (54%). The assay had the highest positivity rate for abscess fluid (77%), abscess tissue (68%), wound biopsies (62%), and heart valves (61%).
Figure 3.
Additional value of targeted metagenomics (tNGS). A, The x-axis is the positivity rate of the test (calculated as the number positive by the tNGS assay divided by the total number of samples in each group on the y-axis). Incremental yield of the tNGS assay offered by NGS was calculated as the number of positive results by NGS divided by the number of samples positive by Sanger sequencing and displayed as + XX% next to each bar group. The “other tissue” group includes biopsies of serous membranes, muscle, organs (brain, liver, spleen, kidney, ovary, breast), digestive tract, masses or tumors not identified as abscesses, thrombi, and deep tissues from pacemaker pockets. The “other fluid” group includes mediastinal fluid, abdominal fluid (including bile), and any fluid not identified as being from an abscess. B, Descriptive microbiology of the microorganisms sequenced by tNGS in the 747 positive samples. Of the 607 monomicrobial samples, 529 (87%) were identified to the species level and 78 (13%) to the genus level only. For formatting reasons, some bacteria are grouped by genus in the figure, even if identified to the species level. Abbreviations: Ct, cycle threshold; NGS, next-generation sequencing.
Microbiology of tNGS-Positive Samples
Nineteen percent of positive samples were polymicrobial. The most common microorganisms detected alone were streptococci (17%), Staphylococcus aureus complex (14%), and Staphylococcus epidermidis (7%). Difficult-to-grow organisms were found, including mycobacteria (4%), Bartonella species (2%), Nocardia species (1%), Ureaplasma species (0.7%), Mycoplasma species (0.5%), and Tropheryma whipplei (0.5%; Figure 3B).
Clinical Analysis
Sample Characteristics
There were 735 Mayo Clinic patients who met inclusion criteria for clinical analysis. A total of 140 duplicates and 16 samples without culture were excluded, leaving 579 for analysis (337 infected and 242 uninfected; Supplementary Figure 1). In the infected group, 60 (18%) had fever at sampling; the medians of C-reactive protein in the infected and uninfected groups were 21.5 and 6.9 g/L, respectively (P < .001). Overall, 269 (46%) patients had received antibiotics in the 4 weeks before sampling, 214 (64%) and 55 (23%) in the infected and uninfected groups, respectively. In the infected group, 134 (40%) had Ct values <32, with 132 (99%) reported positive; 139 (43%) had Ct values >34 with no Tm peak. In the uninfected group, 211 (87%) had Ct values >34, with 3 (1%) having a Ct value <32 (Table 1).
Table 1.
Patient Characteristics at the Time of Sampling
| Characteristic | Total samples n = 579 | Infection n = 337 | No infection n = 242 | |
|---|---|---|---|---|
| n (%) | n (%) | n (%) | P Value | |
| Tissue samples | 281 (49) | 214 (64) | 67 (28) | |
| Fluid samples | 298 (51) | 123 (36) | 175 (72) | |
| Abnormal temperature (>38.5°C or <36°C) | 72 (12) | 60 (18) | 12 (5) | <0.001 |
| Peripheral blood leukocytes (G/L), n = 532 | 7.9 (6.2–10.3) | 8.5 (6.6–11.4) | 7.2 (5.9–9.3) | <0.001 |
| C-reactive protein (g/L), n = 473 | 21.5 (4.5–60.2) | 38.1 (10.2–85.1) | 6.9 (0–23.4) | <0.001 |
| Erythrocyte sedimentation rate (mm/h), n = 426 | 27 (9–55) | 36 (16–73) | 21 (5–40) | <0.001 |
| Gram stain | n = 418 | n = 268 | n = 150 | - |
| Positive Gram stain | 24 (6) | 22 (8) | 2 (0.8) | 0.003 |
|
Conventional cultures
| ||||
| Positive cultures | 141 (24) | 140 (42) | 1 (0.4) | - |
| Negative cultures | 438 (76) | 197 (58) | 241 (99.6) | - |
|
tNGS
| ||||
| Positive | 183 (32) | 178 (53) | 5 (2) | - |
| Negative | 396 (68) | 159 (47) | 237 (98) | - |
| Samples with Ct <32 | 137 (24) | 134 (40) | 3 (1) | - |
| Positive with Sanger sequencing | 105 (77) | 104 (77) | 1 (33) | - |
| Positive with NGS/number sent to NGS | 29/32 (91) | 28/30 (93) | 1/2 (50) | - |
| Samples with Ct ≥32 to ≤34 | 86 (15) | 58 (17) | 28 (12) | - |
| Positive with NGS/number sent to NGS | 45/86 (52) | 42/58 (72) | 3/28 (11) | - |
| Samples with Ct >34 | 356 (61) | 144 (43) | 211 (87) | - |
| Positive with NGS/number sent to NGS | 4/5 (80) | 4/5 (80) | 0/0 | - |
| Antibiotics administered in the 4 weeks before test | 269 (46) | 214 (64) | 55 (23) | - |
| Impact on clinical decision-making | 48 (8) | 46 (14) | 2 (0.8) | - |
| Escalation of treatment | 17 (3) | 17 (5) | 0 (0) | - |
| De-escalation of treatment | 31 (5) | 29 (9) | 2 (0.8) | - |
| Concordant positivity/negativity between tNGS and cultures | 469 (81) | 231 (69) | 238 (98) | - |
| tNGS positive and culture positive | 107 (19) | 106 (31) | 1 (0.4) | - |
| tNGS positive and culture negative | 76 (13) | 72 (21) | 4 (2) | - |
| tNGS negative and culture positive | 34 (6) | 34 (10) | 0 (0) | - |
| tNGS negative and culture negative | 362 (56) | 125 (37) | 237 (98) | - |
Abbreviations: Ct, cycle threshold; NGS, next-generation sequencing; tNGS, targeted metagenomic sequencing–based approach.
Characteristics at the time of sampling were recorded. Medians and 25–75 interquartile ranges are displayed for quantitative values. Numbers and percentages are displayed for qualitative values.
Impact on Clinical Decision-Making
The tNGS test impacted clinical decision-making in 48 (8%) cases, 46 (14%) and 2 (0.8%) in the infected and uninfected groups, respectively. Treatment de-escalation was the most common impact, occurring in 31 (5%) overall, including 2 (1%) in the uninfected group. Treatment was escalated in 17 (3%), all of whom were infected.
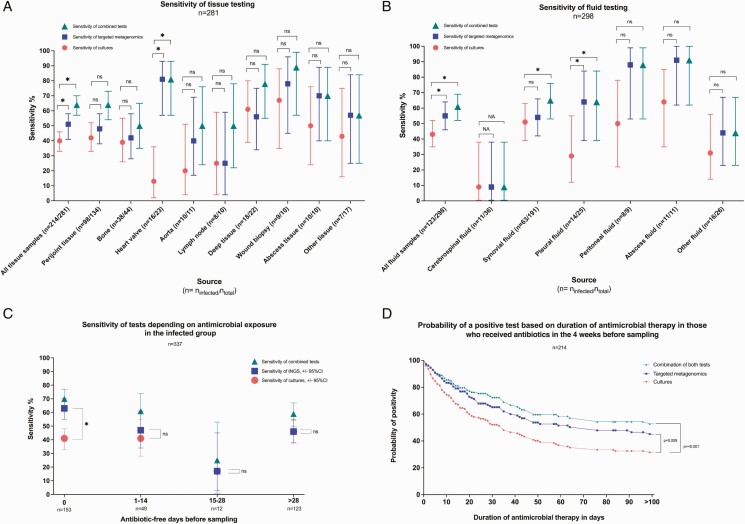
Clinical sensitivity of culture was 42% overall and 40% and 43% in the tissue and fluid groups, respectively. tNGS was more sensitive than culture overall (53%, P < .001), in the tissue group (51%, P < .001), the fluid group (55%, P < .001), and the group that received antimicrobial therapy in the 4 weeks prior to sampling (57%, P < .001; Table 2, Figure 4A, 4B). Clinical sensitivity of culture (46%) and tNGS (46%) were comparable (P = .9) in the group that had not received antimicrobics in the 4 weeks before sampling; the negative predictive value in this group was 74% for culture and tNGS and 78% with both tests combined. Consideration of both methods together increased clinical sensitivity to 59% overall, with 97% specificity. The increased yield of tNGS over culture was especially marked in patients receiving antibiotics at sampling (sensitivity: 41% for culture, 63% for tNGS, and 70% for the combination thereof; Figure 4C). Differential sensitivity between the 2 tests generally decreased as the number of antibiotic-free days before sampling increased. The probability of a positive test was higher with tNGS compared with culture over the duration of antimicrobial therapy exposure (P = .009; Figure 4D). In the infected group, 72 (37%) culture-negative samples were positive by tNGS (Supplementary Figure 2C).
Table 2.
Clinical Analysis of the Targeted Metagenomics Sequencing-based Approach and Cultures
| Sensitivity, % (95% CI interval) |
Specificity, % (95% CI) |
Negative Predictive Value, % (95% CI) |
Positive Predictive Value, % (95% CI) |
|
|---|---|---|---|---|
| All samples, n = 579 | ||||
| Cultures | 42 (37–47) | 100 (98–100) | 55 (50–60) | 99 (96–100) |
| Targeted metagenomics | 53a (47–58) | 98 (95–99) | 60 (55–65) | 97 (94–99) |
| Cultures and/or targeted metagenomics | 63 a (58–68) | 98 (95–99) | 65 (60–70) | 98 (95–99) |
| Tissue samples, n = 281 | ||||
| Cultures | 40 (33–46) | 99 (92–100) | 34 (28–41) | 99 (94–100) |
| Targeted metagenomics | 51 a (45–58) | 97 (90–99) | 38 (31–46) | 98 (94–100) |
| Cultures and/or targeted metagenomics | 64 a (57–70) | 97 (90–99) | 46 (38–54) | 99 (95–100) |
| Fluid samples, n = 298 | ||||
| Cultures | 43 (35–52) | 100 (98–100) | 71 (65–77) | 100 (93–100) |
| Targeted metagenomics | 55 a (46–64) | 98 (95–100) | 76 (70–81) | 96 (88–99) |
| Cultures and/or targeted metagenomics | 61 a (52–69) | 98 (95–100) | 78 (72–83) | 96 (89–99) |
| Antibiotics received administered in the 4 weeks before sampling, n = 269 | ||||
| Cultures | 39 (33–46) | 98 (90–100) | 29 (23–36) | 99 (94–100) |
| Targeted metagenomics | 57 a (50–63) | 95 (85–99) | 36 (29–44) | 98 (93–99) |
| Cultures and/or targeted metagenomics | 65 a (59–71) | 95 (85–99) | 41 (33–50) | 98 (94–99) |
| No antibiotics received administered in the 4 weeks before sampling, n = 310 | ||||
| Cultures | 46 (37–54) | 100 (98-100) | 74 (68–79) | 100 (94–100) |
| Targeted metagenomics | 46 (38–55) | 99 (96–100) | 74 (68–79) | 97 (88–99) |
| Cultures and/or targeted metagenomics | 59 a (50–67) | 99 (96–100) | 78 (73–83) | 97 (91–100) |
Abbreviation: CI, confidence interval.
P < .001 compared to cultures.
Figure 4.
Clinical sensitivity by sample type and antimicrobial exposure. A, Clinical sensitivity (determined as the percentage of positive identifications in the infected group) of cultures, tNGS, and the combination of the 2 for tissue samples by source. B, Clinical sensitivity (percentage of positive identifications in the infected group) of cultures, tNGS, and the combination of the 2 for fluid samples by source. C, Sensitivity (percentage of positive identifications in the infected group) by numbers of days without antimicrobial therapy prior to sampling. Sensitivities of cultures and tNGS were compared using the McNemar test of paired proportions. *P value < .05. D, Kaplan-Meier curve showing probability of positivity for cultures, tNGS, and both based on duration of antimicrobial therapy before sampling in the infected group (n = 214). Patients who had received more than 100 days of antimicrobial therapy prior to sampling were included in the analysis but were censored at 100 days on the graphic representation. The probability of culture positivity was compared with tNGS and the combination of the 2 using a log-rank test (Mantel-Cox). Abbreviations: CI, confidence interval; ns = nonsignificant; tNGS, targeted metagenomics.
Concordance
Results (positive or negative) were concordant between culture and tNGS in 81% of cases. In the infected group, discordance between tNGS and cultures was found in 106 samples (31%), with 72 being tNGS-positive/culture-negative and 34 being tNGS-negative/culture-positive (Supplementary Figure 2). A total of 125 samples in the infected group (37%) were both tNGS- and culture-negative, among which 96 (28%) yielded no microorganism by any testing performed (Supplementary Figure 2B, 2C).
DISCUSSION
For the past decade, metagenomic testing for infectious diseases has generally been reserved for research and challenging clinical cases, mainly because of cost and technical difficulty but also because of the uncertainty as to clinical value. This real-life study provides insight as to how tNGS implemented in a high-throughput clinical molecular microbiology laboratory yields benefits in terms of pathogen detection in infection diagnosis and impacts patient care. Addition of NGS increased 16S rRNA gene PCR positivity by 87% compared with Sanger sequencing alone. tNGS positivity was 35% higher than the 13% (78 of 607) and 11% (35 of 312) positivity of Sanger sequencing alone that were reported by Aggarwal et al [10] and Yoo et al [20], respectively. In addition, tNGS identified multiple difficult-to-grow organisms, confirming its use for infection types associated with delayed diagnoses [21].
Some limitations of Sanger sequencing were circumvented with NGS, in particular, detection of polymicrobial samples (which represented 19% of positive samples in this study). For sample types with a high likelihood of polymicrobial results, such as lung tissue, peritoneal fluid, wound samples, and pleural fluid, Sanger sequencing sometimes failed in more than half of cases, even with low Ct values, requiring NGS such that direct NGS regardless of the Ct value deserves further study. Advantages of using NGS for such specimen types have been highlighted by others [16, 22–24].
This study is the first, to date, to describe the clinical performance of a tNGS approach using a large number of samples, with results showing that tNGS has higher clinical sensitivity than cultures (albeit in a study population likely biased to culture negativity), with high specificity (98%) and positive and negative predictive values of 97% and 60%, respectively.
An alternative to tNGS is sNGS. One study evaluated sNGS in a clinical setting using 109 samples of several types and reported a sensitivity and specificity of 67% and 69%, respectively [25]. Others studied sNGS on plasma and lower respiratory tract specimens and reported high sensitivities (>90%), albeit with low specificities of 63% [26] and 42% [27], respectively. False-positive results have possible consequences in terms of overprescription of antimicrobial agents and missed/delayed diagnosis of “real” diagnoses. Gu et al evaluated sNGS to detect bacteria in body fluids and reported a sensitivity of 79% and specificity of 91%, but only on samples with a confirmed diagnosis by culture or PCR [28]. In our cohort, we calculated clinical sensitivity, with 197 culture negative samples, including 96 that yielded no pathogen identification by any other mean. sNGS is, however, theoretically better suited than 16S rRNA gene–based tNGS for infections where viruses and/or fungi are dominant, such as meningitis, meningoencephalitis, and eye infections [29, 30]. Addition of a conserved fungal gene into a tNGS approach may be helpful.
tNGS had an impact on clinical decision-making for 8% of samples overall, 14% in the infected group and 0.8% in the uninfected group. Clinicians might have been cautious about changing management since this was a new assay. These numbers are comparable to those from a French study on 16S rRNA PCR/Sanger sequencing where an impact on clinical decisions was found in 62 of 806 (8%) overall and 62 of 109 (32%) positive samples [31]. The effect of antimicrobial therapy on tNGS was also evaluated. tNGS was more sensitive than culture among patients who received antimicrobial treatment at sampling. This difference decreased over time as the number of antibiotic-free days increased. Clinical sensitivity of tNGS was higher than culture among patients with prolonged antibiotic exposure.
There are limitations of this study. In the infected group, 43% of samples were not sequenced because they had a Ct >34. Use of Ct values to drive the sequencing trajectory was intentional to avoid sequencing samples with a low chance of positivity, reducing labor, cost, and the potential for false-positive results. In previous research, a threshold Ct of 34 was proposed to differentiate positive and negative samples, with specific application to arthroplasty-derived sonicate fluids [15]. This cutoff might not apply equally to all sample types and therefore should be evaluated by sample source and infection type. That there were diverse sample types in this study is also a limitation. The definition of infection was based on assessment of the clinical team. Despite being imperfect, this represents real life and has been previously used to assess clinical sensitivity of metagenomic tests in infectious diseases [32]. Providers were not blinded to results of tNGS. Consequently, for patients with negative cultures and doubtful presence of infection, tNGS negativity may have made providers more likely to observe patients off antibiotics. Clinical sensitivity was calculated because the aim was to correlate the presence of infection with results of the assay; hence, the low sensitivity. Interpretation of results requires a trained expert who can adjudicate results and understand assay background. Cutibacterium acnes and S. epidermidis, which may be contaminants or pathogens, were not typically reported as pathogens if found in negative controls. While a cost analysis was not performed, the cost of tNGS is several-fold less than that of sNGS [33, 34]. Theoretical turnaround time is dependent on Ct value, with a result in less than 24 hours for negative samples or those sent to Sanger sequencing, and 36–48 hours (ideally) for samples sent to NGS. However, maintaining a 48-hour turnaround time for NGS in clinical practice is challenging, with time to delivery of results being as long as 10 days in some cases. To improve time to results, trained technologists would need to be available 24/7 to perform NGS. Nevertheless, turnaround time of cultures for certain organisms such as mycobacteria may be longer than 10 days tNGS can still be clinically relevant. Performance of the test on FFPE compared with fresh tissue, which is a controversial subject, was not assessed [35, 36]. Finally, because a 16S rRNA PCR approach was used, prediction of antibiotic resistance was not possible. The tNGS test that was studied was only used for normally sterile human tissues and body fluids. Sampling of plasma or whole blood, particularly for acutely ill patients or those for whom the site of infection is not known [37], or for diagnosis of tick-borne pathogens, immunocompromised patients, or septic patients [38–41], or sampling of other specimens, such as respiratory secretions or skin, requires further study.
In conclusion, results of this study show the benefit of adding NGS to a Sanger sequencing–based 16S rRNA PCR assay and overall value of tNGS in a clinical setting. This approach is not meant to replace culture but to serve as a complementary diagnostic tool for fastidious bacteria, patients who receive antibiotics before sampling, and/or to tease apart undefined etiologies of potential infections.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health (NIH).
Financial support. Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the NIH under R01AR05664.
Contributor Information
Laure Flurin, Division of Clinical Microbiology, Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, Minnesota, USA; Department of Intensive Care, University Hospital of Guadeloupe, Pointe-à-Pitre, France.
Matthew J Wolf, Division of Clinical Microbiology, Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, Minnesota, USA.
Melissa M Mutchler, Division of Clinical Microbiology, Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, Minnesota, USA.
Matthew L Daniels, Division of Clinical Microbiology, Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, Minnesota, USA.
Nancy L Wengenack, Division of Clinical Microbiology, Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, Minnesota, USA.
Robin Patel, Division of Clinical Microbiology, Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, Minnesota, USA; Division of Infectious Diseases, Department of Medicine, Mayo Clinic, Rochester, Minnesota, USA.
References
- 1. Lefterova MI, Suarez CJ, Banaei N, Pinsky BA.. Next-generation sequencing for infectious disease diagnosis and management: a report of the Association for Molecular Pathology. J Mol Diagn 2015; 17:623–34. [DOI] [PubMed] [Google Scholar]
- 2. Sigakis MJG, Jewell E, Maile MD, Cinti SK, Bateman BT, Engoren M.. Culture-negative and culture-positive sepsis: a comparison of characteristics and outcomes. Anesth Analg 2019; 129:1300–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fournier PE, Thuny F, Richet H, et al. . Comprehensive diagnostic strategy for blood culture-negative endocarditis: a prospective study of 819 new cases. Clin Infect Dis 2010; 51:131–40. [DOI] [PubMed] [Google Scholar]
- 4. Harbarth S, Nobre V, Pittet D.. Does antibiotic selection impact patient outcome?. Clin Infect Dis 2007; 44:87–93. [DOI] [PubMed] [Google Scholar]
- 5. Spyridakis E, Gerber JS, Schriver E, et al. . Clinical features and outcomes of children with culture-negative septic arthritis. J Pediatric Infect Dis Soc 2019; 8:228–34. [DOI] [PubMed] [Google Scholar]
- 6. Teshome BF, Vouri SM, Hampton N, Kollef MH, Micek ST.. Duration of exposure to antipseudomonal beta-lactam antibiotics in the critically ill and development of new resistance. Pharmacotherapy 2019; 39:261–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Durand ML. Bacterial and fungal endophthalmitis. Clin Microbiol Rev 2017; 30:597–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gupta S, Sakhuja A, Kumar G, McGrath E, Nanchal RS, Kashani KB.. Culture-negative severe sepsis: nationwide trends and outcomes. Chest 2016; 150:1251–9. [DOI] [PubMed] [Google Scholar]
- 9. Nitzan O, Brodsky Y, Edelstein H, et al. . Microbiologic data in acute cholecystitis: ten years’ experience from bile cultures obtained during percutaneous cholecystostomy. Surg Infect (Larchmt) 2017; 18:345–9. [DOI] [PubMed] [Google Scholar]
- 10. Aggarwal D, Kanitkar T, Narouz M, Azadian BS, Moore LSP, Mughal N.. Clinical utility and cost-effectiveness of bacterial 16S rRNA and targeted PCR based diagnostic testing in a UK microbiology laboratory network. Sci Rep 2020; 10:7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fida M, Khalil S, Abu Saleh O, et al. . Diagnostic value of 16S ribosomal RNA gene polymerase chain reaction/Sanger sequencing in clinical practice. Clin Infect Dis 2021; 73:961–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gomez E, Cazanave C, Cunningham SA, et al. . Prosthetic joint infection diagnosis using broad-range PCR of biofilms dislodged from knee and hip arthroplasty surfaces using sonication. J Clin Microbiol 2012; 50:3501–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Behjati S, Tarpey PS.. What is next generation sequencing?. Arch Dis Child Educ Pract Ed 2013; 98:236–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dyrhovden R, Ovrebo KK, Nordahl MV, Nygaard RM, Ulvestad E, Kommedal O.. Bacteria and fungi in acute cholecystitis. A prospective study comparing next generation sequencing to culture. J Infect 2020; 80:16–23. [DOI] [PubMed] [Google Scholar]
- 15. Flurin L, Wolf MJ, Greenwood-Quaintance KE, Sanchez-Sotelo J, Patel R.. Targeted next generation sequencing for elbow periprosthetic joint infection diagnosis. Diagn Microbiol Infect Dis 2021; 101:115448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stebner A, Ensser A, Geissdorfer W, Bozhkov Y, Lang R.. Molecular diagnosis of polymicrobial brain abscesses with 16S-rDNA-based next-generation sequencing. Clin Microbiol Infect 2021; 27:76–82. [DOI] [PubMed] [Google Scholar]
- 17. Kommedal O, Simmon K, Karaca D, Langeland N, Wiker HG.. Dual priming oligonucleotides for broad-range amplification of the bacterial 16S rRNA gene directly from human clinical specimens. J Clin Microbiol 2012; 50:1289–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Osmon DR, Berbari EF, Berendt AR, et al. . Diagnosis and management of prosthetic joint infection: clinical practice guidelines by the Infectious Diseases Society of America. Clin Infect Dis 2013; 56:e1–e25. [DOI] [PubMed] [Google Scholar]
- 19. Peel TN, Sedarski JA, Dylla BL, et al. . Laboratory workflow analysis of culture of periprosthetic tissues in blood culture bottles. J Clin Microbiol 2017; 55:2817–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yoo IY, Kang OK, Lee MK, et al. . Comparison of 16S ribosomal RNA targeted sequencing and culture for bacterial identification in normally sterile body fluid samples: report of a 10-year clinical laboratory review. Ann Lab Med 2020; 40:63–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Van Scoy RE. Culture-negative endocarditis. Mayo Clin Proc 1982; 57:149–54. [PubMed] [Google Scholar]
- 22. Dyrhovden R, Nygaard RM, Patel R, Ulvestad E, Kommedal O.. The bacterial aetiology of pleural empyema. A descriptive and comparative metagenomic study. Clin Microbiol Infect 2019; 25:981–6. [DOI] [PubMed] [Google Scholar]
- 23. Shiraishi Y, Kryukov K, Tomomatsu K, et al. . Diagnosis of pleural empyema/parapneumonic effusion by next-generation sequencing. Infect Dis (Lond) 2021; 53:450–9. [DOI] [PubMed] [Google Scholar]
- 24. Salipante SJ, Sengupta DJ, Rosenthal C, et al. . Rapid 16S rRNA next-generation sequencing of polymicrobial clinical samples for diagnosis of complex bacterial infections. PLoS One 2013; 8:e65226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang HC, Ai JW, Cui P, et al. . Incremental value of metagenomic next generation sequencing for the diagnosis of suspected focal infection in adults. J Infect 2019; 79:419–25. [DOI] [PubMed] [Google Scholar]
- 26. Blauwkamp TA, Thair S, Rosen MJ, et al. . Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat Microbiol 2019; 4:663–74. [DOI] [PubMed] [Google Scholar]
- 27. Charalampous T, Kay GL, Richardson H, et al. . Nanopore metagenomics enables rapid clinical diagnosis of bacterial lower respiratory infection. Nat Biotechnol 2019; 37:783–92. [DOI] [PubMed] [Google Scholar]
- 28. Gu W, Deng X, Lee M, et al. . Rapid pathogen detection by metagenomic next-generation sequencing of infected body fluids. Nat Med 2021; 27:115–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kuo MT, Chen JL, Hsu SL, Chen A, You HL.. An omics approach to diagnosing or investigating fungal keratitis. Int J Mol Sci 2019; 20:3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wilson MR, Sample HA, Zorn KC, et al. . Clinical metagenomic sequencing for diagnosis of meningitis and encephalitis. N Engl J Med 2019; 380:2327–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ursenbach A, Schramm F, Severac F, et al. . Revised version (INFD-D-20-00242): impact of 16S rDNA sequencing on clinical treatment decisions: a single center retrospective study. BMC Infect Dis 2021; 21:190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee RA, Al Dhaheri F, Pollock NR, Sharma TS.. Assessment of the clinical utility of plasma metagenomic next-generation sequencing in a pediatric hospital population. J Clin Microbiol 2020; 58:e00419-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Akram A, Maley M, Gosbell I, Nguyen T, Chavada R.. Utility of 16S rRNA PCR performed on clinical specimens in patient management. Int J Infect Dis 2017; 57:144–9. [DOI] [PubMed] [Google Scholar]
- 34. Govender KN, Street TL, Sanderson ND, Eyre DW.. Metagenomic sequencing as a pathogen-agnostic clinical diagnostic tool for infectious diseases: a systematic review and meta-analysis of diagnostic test accuracy studies. J Clin Microbiol 2021; 59:e0291620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gao XH, Li J, Gong HF, et al. . Comparison of fresh frozen tissue with formalin-fixed paraffin-embedded tissue for mutation analysis using a multi-gene panel in patients with colorectal cancer. Front Oncol 2020; 10:310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hedegaard J, Thorsen K, Lund MK, et al. . Next-generation sequencing of RNA and DNA isolated from paired fresh-frozen and formalin-fixed paraffin-embedded samples of human cancer and normal tissue. PLoS One 2014; 9:e98187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Grumaz S, Stevens P, Grumaz C, et al. . Next-generation sequencing diagnostics of bacteremia in septic patients. Genome Med 2016; 8:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fida M, Wolf MJ, Hamdi A, et al. . Detection of pathogenic bacteria from septic patients using 16S ribosomal RNA gene-targeted metagenomic sequencing. Clin Infect Dis 2021; 73:1165–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Long Y, Zhang Y, Gong Y, et al. . Diagnosis of sepsis with cell-free DNA by next-generation sequencing technology in ICU patients. Arch Med Res 2016; 47:365–71. [DOI] [PubMed] [Google Scholar]
- 40. Thoendel M. Targeted metagenomics offers insights into potential tick-borne pathogens. J Clin Microbiol 2020; 58:e01893-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vijayvargiya P, Jeraldo PR, Thoendel MJ, et al. . Application of metagenomic shotgun sequencing to detect vector-borne pathogens in clinical blood samples. PLoS One 2019; 14:e0222915. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.