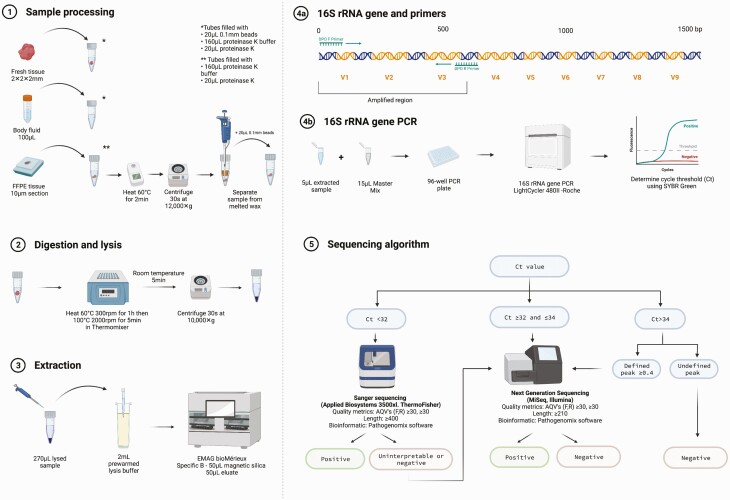
Figure 1.
Laboratory workflow of the targeted metagenomics approach (tNGS). (1) A 2-mm3 piece of tissue or 100 µL of fluid was placed into a lysis tube with 20 µL of 0.1-mm silica/zirconium beads, 160 µL proteinase K buffer (PKB), and 20 µL of proteinase K. For FFPE tissues, a 10-µm section was put into a 1.5-mL centrifuge tube with 160 µL of PKB and 20 µL of proteinase K and heated to 60°C for 2 minutes to melt the wax. After centrifugation, liquid and tissue below the wax were transferred to a lysis tube with 20 µL of 0.1-mm beads. (2) Lysis tubes were incubated/spun, cooled for 5 minutes at room temperature, and then centrifuged. (3) Then, 2 mL of prewarmed (40°C) NUCLISENS easyMAG lysis buffer was added to the disposable NUCLISENS easyMAG cartridge with a maximum of 270 µL of the lysed sample and incubated at room temperature for 10 minutes followed by extraction loading. (4) Next, the 16S rRNA gene polymerase chain reaction targeted the V1–V3 region of the bacterial 16S rRNA gene using dual priming oligonucleotides. (5) Depending on the Ct value, samples were sent to Sanger or next-generation sequencing or reported negative. Abbreviations: AQV, average quality value; Ct, cycle threshold; FFPE, formalin-fixed paraffin-embedded; F,R, forward, reverse; PCR, polymerase chain reaction. Created with Biorender.com.