Figure 2. Validation of conditional endothelial-specific Tfrc knockout mice.
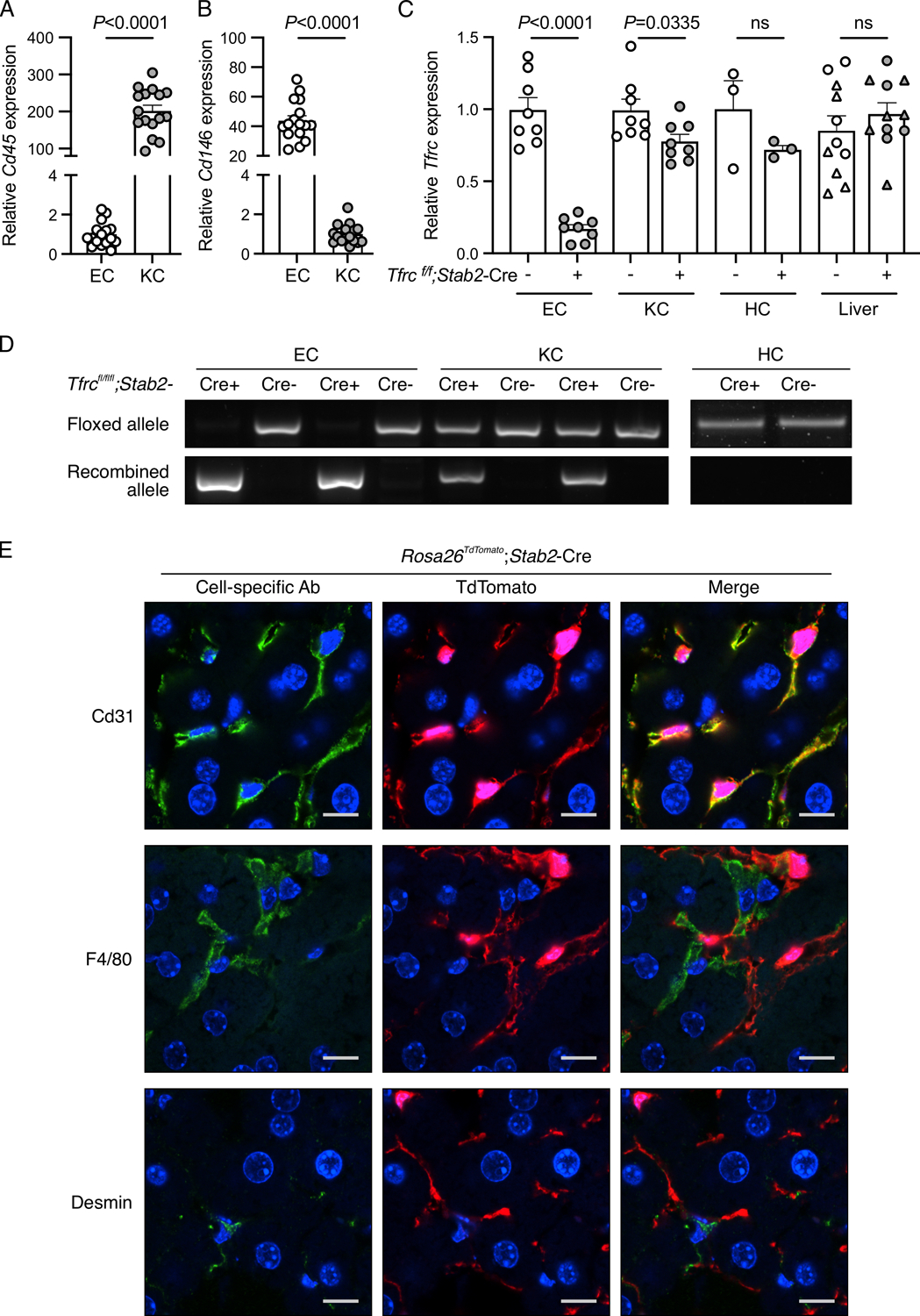
(A-D) Liver endothelial cells (EC) and Kupffer cells (KC) isolated by fluorescence-activated cell sorting, hepatocytes (HC) isolated by density-gradient centrifugation, and total livers from 8–16-week-old Tfrcfl/fl;Stab2-Cre+ and Cre− mice were analyzed for: (A-B) expression of Cd45 and Cd146 relative to Rpl19 by qRT-PCR to assess EC and KC purity; (C) Tfrc relative to Rpl19 by qRT-PCR; and (D) floxed and recombined Tfrc alleles by PCR. For panels (A-B), cells from Cre- and Cre+ mice were combined for analysis. For panel (C), whole liver analysis includes male (circles) and female (triangles) mice. For panel (D), representative gel images from N=8/group (EC, KC) and N=3/group (HC) are shown. Graphs represent mean ± SEM with individual points indicating the number of animals per group. Statistical differences were determined by two-tailed Student’s t-test. (E) Confocal immunofluorescence microscopy images depicting colocalization of cell-specific markers with tdTomato in livers from Rosa26Sortm9-tdTomato;Stab2-Cre+ reporter mice. Liver sections were stained for specific cell populations using CD31 (endothelial cells), F4/80 (Kupffer cells), or desmin (stellate cells) antibodies (green). Cells with Cre recombinase activity are endogenously red because of the removal of a stop codon upstream of the tdTomato fluorescent protein. Nuclei are labeled blue with 4′,6-diamidino-2-phenylindole. Scale bars represent 10 μm. Representative images from N=3 mice/group are shown.
