Figure 2.
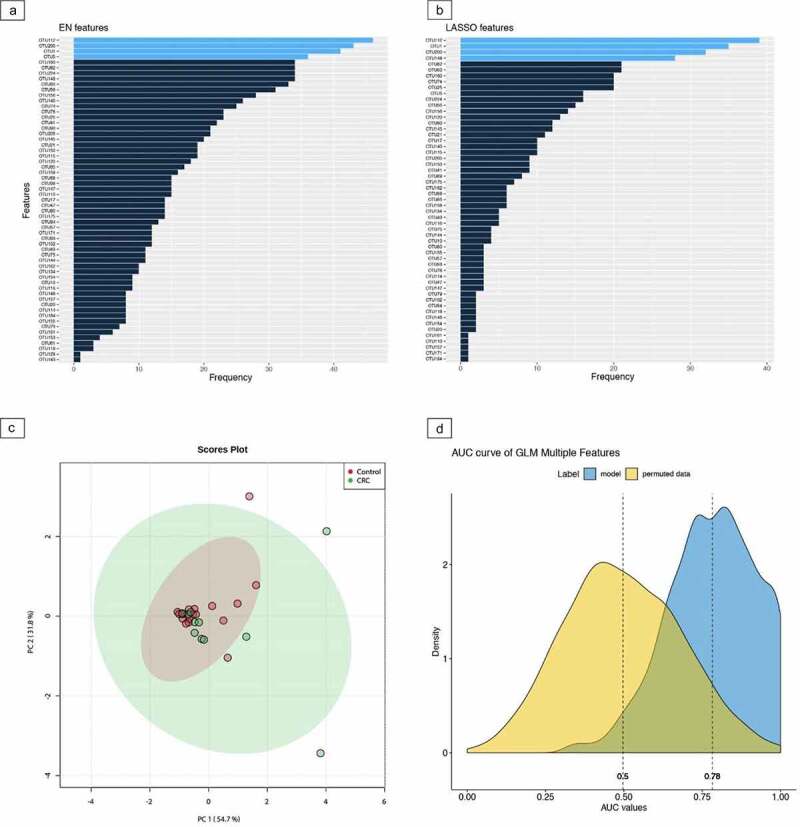
Machine learning pipeline for colorectal cancer and controls using microbial taxa The entire machine learning pipeline for the comparison between fecal samples of colorectal cancer and controls based on microbial taxa. part a and b depict the outcomes of the elastic net (EN) and least absolute shrinkage and selection operator (LASSO) feature selection methods, respectively. The light blue color in both methods indicates the first quartile of the ranked features across 100 iterations. The 5 selected markers are Methanobrevibacter, Bifidobacterium, Eubacterium hallii, Ruminococcaceae UCG-003 and Desulfovibrio. In part c, the relatedness of the selected markers is depicted using principal component analysis (PCA). Part d depicts the stability plot obtained with logistic regression models for the combined marker panel that has been selected. Corresponding area under the curve (AUC) is presented in blue.
