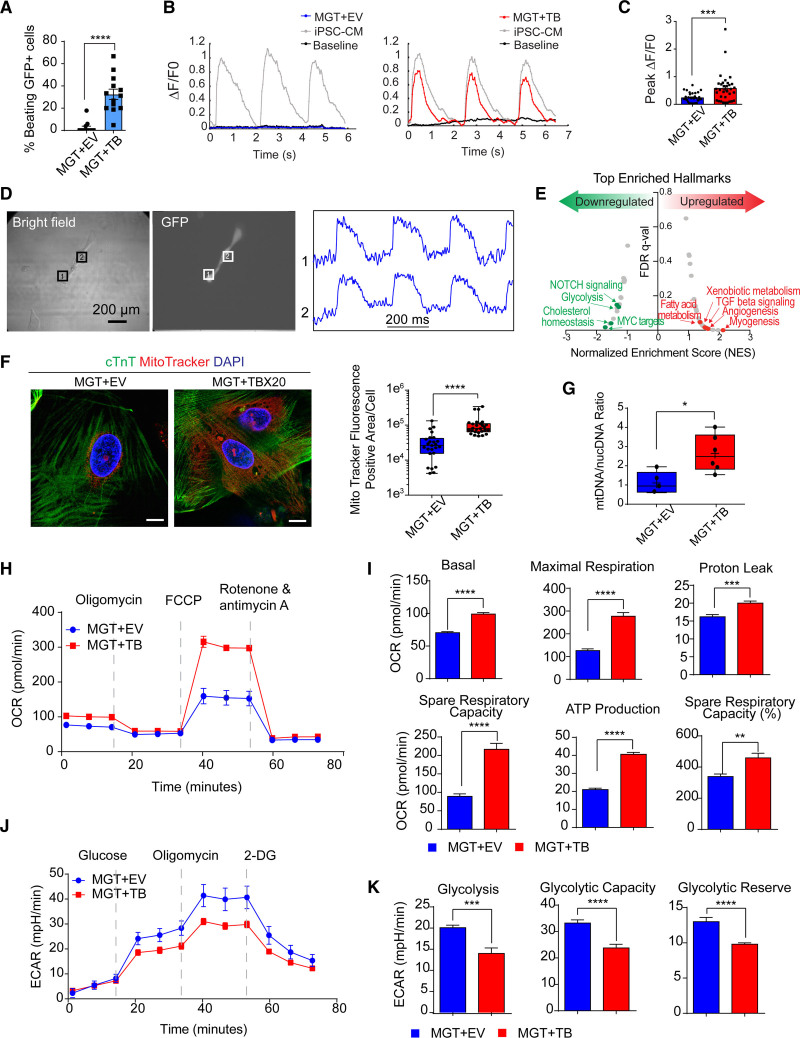
Figure 4.
TBX20 overexpression leads to functional improvement of hiCMs. A, Quantification of observed beating human induced cardiomyocytes (hiCMs) derived from H9-derived fibroblasts (H9Fs) after 1 month of co-culture with human induced pluripotent stem cell–derived cardiomyocytes (iPSC-CMs; n=10 for each group, repeated 2 times independently). B, Tracing and measurements from microscopy imaging show calcium transients in MGT + empty vector (EV) or MGT+TBX20 co-cultured cells as indicated in A. C, Quantification of measured peak ΔF/F0 in co-cultured hiCMs (n=33, repeated 2 times independently). D, Bright field and GFP (green fluorescent protein) fluorescence images showing MGT+TBX20-induced individual hiCMs after co-culture. Right panel shows the selected optical traces of membrane potential (Vm) during spontaneous beating with a cycle length of 270 ms. E, Gene set enrichment analysis of RNA sequencing data from H9F-derived hiCMs showing the enriched hallmarks in TBX20 upregulated and downregulated genes. F, Representative images and quantification of mitochondria content labeled using MitoTracker in cardiac troponin T (cTnT)+ hiCMs derived from H9F (n=20 per group). Scale bars, 10 μm. G, Reverse transcription quantitative polymerase chain reaction results showing the relative ratio of mitochondria DNA to the total genomic DNA in H9F-derived hiCMs (n=6 per group). H, Representative oxygen consumption rate profile analyzed from mito-stress Seahorse assay in hiCMs derived from H9Fs (n=10 per group, repeated 3 times independently). I, Quantification of basal respiration, maximal respiration, proton leak, spare respiratory capacity, adenosine triphosphate (ATP) production, and spare respiratory capacity as a percentage in hiCMs treated with MGT+TBX20 or MGT+EV from H9Fs (n=10 per group). J, Representative extracellular acidification rate profile analyzed from glucose-stress Seahorse assay in TBX20-treated or EV-treated hiCMs derived from H9Fs (n=10 per group). K, Quantification of glycolysis, glycolytic capacity, and glycolytic reserve in H9F-derived hiCMs transduced with TBX20 or EV (n=10 per group). All data are expressed as mean ± SEM (A, C, F, G, I, and K, Student t test). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. 2-DG indicates 2-deoxy-glucose; ECAR‚ extracellular acidification rate; FCCP‚ fluoro-carbonyl cyanide phenylhydrazone; FDR, false discovery rate; and OCR, oxygen consumption rate.