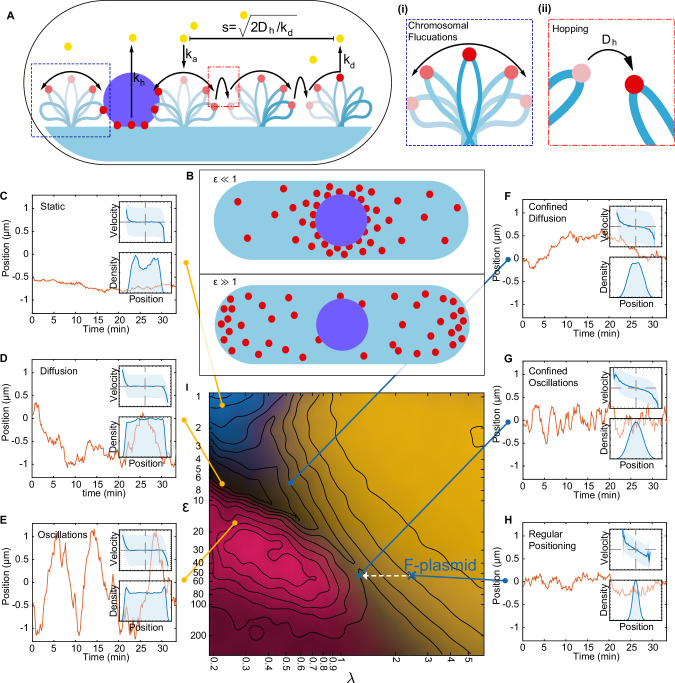
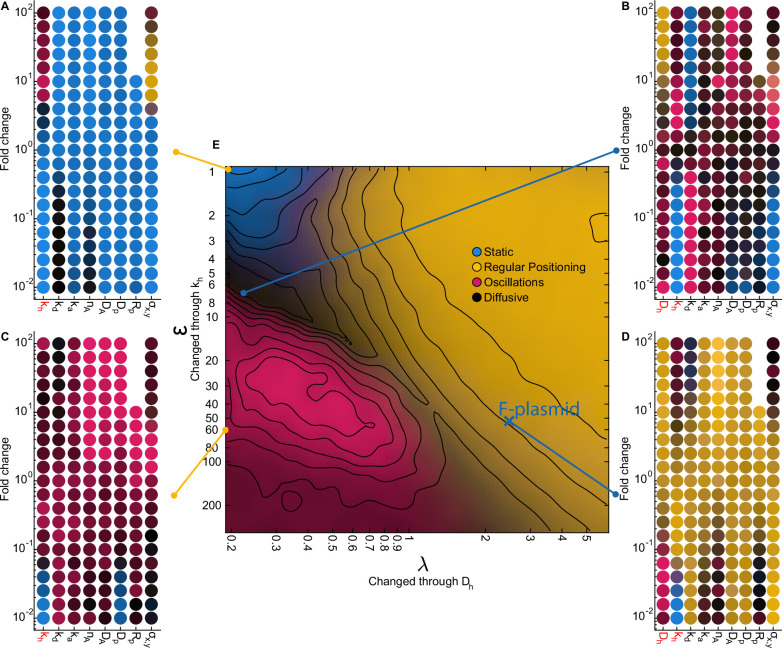
(
A–D) Each scatter plot contains multiple 1D sweeps centred at the corresponding location in our phase diagram. Each 1D sweep (at its extreme) increases or decreases one parameter by a factor of 100 (fold change 10
–2 - 10
2). At a fold change of 10
0 the parameters are the same as at the indicated location. Labels in red indicate parameters which were used to change
and
in our phase diagram (10 simulations per point). The locations for (
A) and (
C) were chosen such that
(no diffusion on the nucleoid). (
D) is located at our predicted parameters for F plasmid. The radius of the plasmid (
) could not be increased more than 10-fold since above that threshold the diameter of the plasmid was greater than the width of the cell. The colours in the scatter plot indicate the behaviour of the system as in
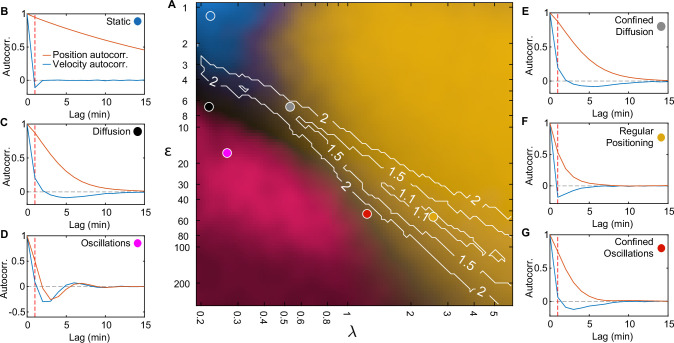
Figure 3I. (
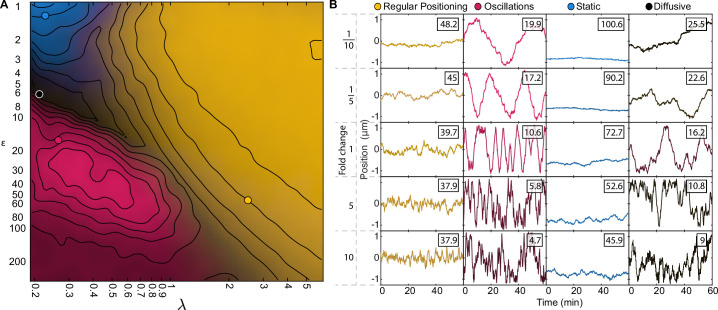
E) Same as
Figure 3I. Notable transitions. (1) Decreasing the tether hydrolysis rate
results in longer lived tethers and hence slower plasmid movement. Beyond a point, the plasmid appears static on the timescale of the simulation. However, we have confirmed by performing longer simulations that it is not static but diffusive for
and regularly positioned for
(as explained in the text, the blue region in the top left of the phase diagram is technically diffusive) as predicted by flux balance. As
is increased in the
regime, an oscillatory transition occurs when the plasmid begins to move faster than hydrolyzed ParA dimers can be replaced resulting in a depletion zone behind the plasmid and directed movement. (2) Decreasing
decreases the diffusive length-scale
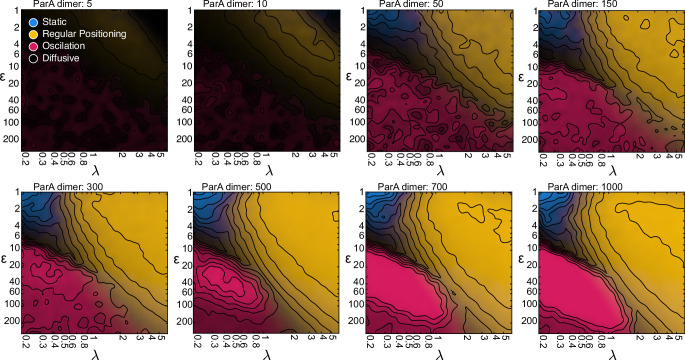
, moving the system out of the regular positioning regime and towards either oscillatory or diffusive dynamics. It also decreases the total flux of ParA into the plasmid leading to fewer tethers but this is not responsible for the nature of the dynamics as increasing
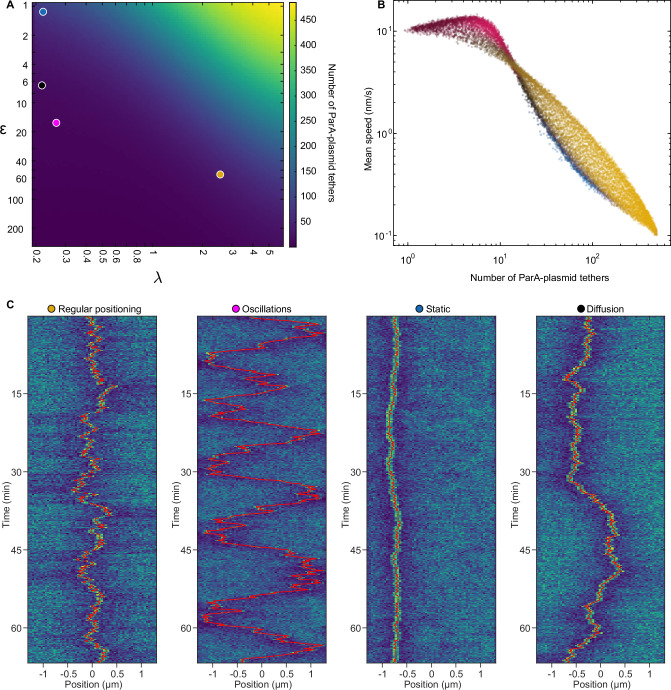
, the total amount of ParA, does not affect the nature of the dynamics (see also
Figure 3—figure supplement 3). (3) Increasing
decreases both
and
(as well as the fraction of nucleoid-bound ParA dimers
) and so moves the system approximately diagonally in the phase diagram. (4) The plasmid diffusion coefficient
is most relevant in the oscillatory regime, in which there are the fewest tethers. Oscillations rely on the plasmid moving faster than hydrolyzed ParA tethers can be replaced. Thus increasing plasmid mobility through
results in stronger directed movement and hence oscillations, while decreasing it moves the system towards more diffusive behaviour (
C). (5) An additional requirement for non-diffusive dynamics is that the tether lifetime is longer that the timescale of the tether-induced ‘pulling’ (
for a single tether). This effect explains the darkening in the phase diagram at the bottom of the oscillatory regime. The same transition to diffusive dynamics occurs at high values of the spring constant
. Note however that for the longest tether lifetime and high
, regular positioned was observed at
(
; no diffusion on the nucleoid) i.e. outside of our claimed regular positioning regime (
A). This occurs because at this unphysical value,
is comparable to the size of the cell and therefore DNA-bound ParA dimers can interact with the plasmid from every location within the cell. The plasmid is therefore positioned at mid-cell because this is where the net force from all tethered dimers balances. In other words geometry sensing occurs, not through the local detection of a disparity in incoming fluxes but through the global detection of all ParA dimers.