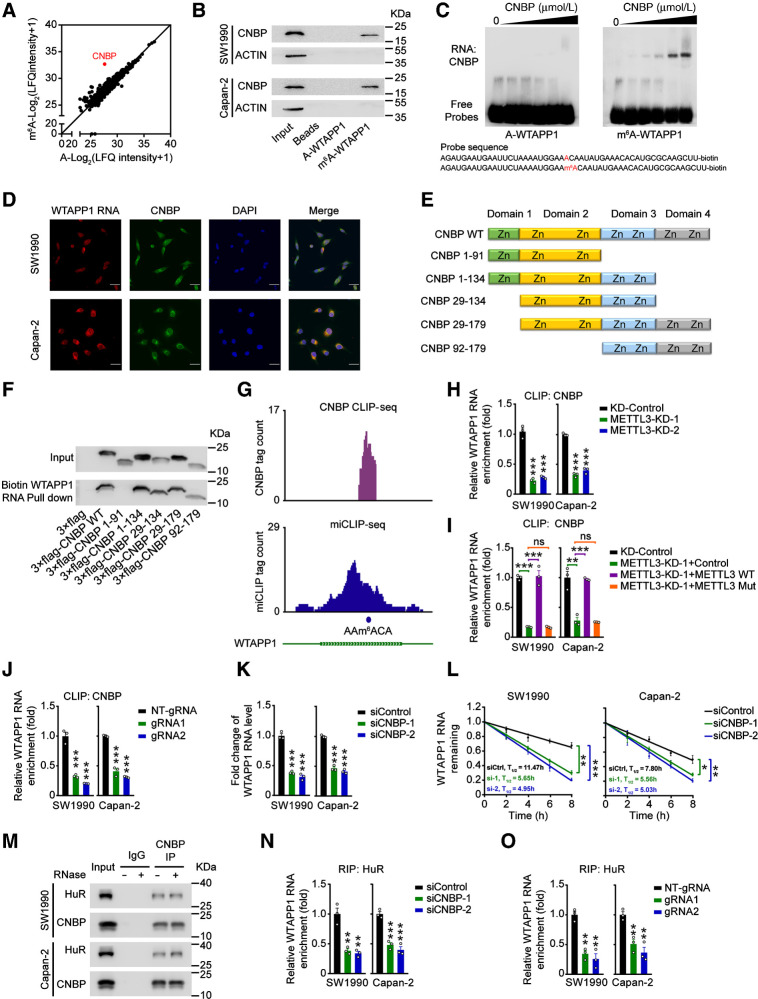
Figure 4.
CNBP is a m6A mediator that stabilizes WTAPP1 RNA. A, Scatter plot of proteins in SW1990 and Capan-2 cells that bound to m6A-modified or nonmodified WTAPP1 probe, both of which were 50-bp oligonucleotides centering on WTAPP1 RNA m6A site. Unique peptide of ≥ 5 was selected as filter criterium. The diagonal line marks the middle values of x and y axes. The red point represents CNBP protein. B, Western blot analysis showed specific association of m6A-WTAPP1 RNA with CNBP identified by proteomic screening. C, Electrophoretic mobility shift assays of recombinant CNBP with m6A-modified or unmodified WTAPP1 RNA probes. The reactions were carried out with a constant level of probes and various amount of recombinant CNBP (0–10 μmol/L). D, Colocalization of WTAPP1 RNA and CNBP protein in cells shown by IF. Scale bars, 30 μm. E and F, Pull-down assays with in vitro–transcribed biotin-labeled WTAPP1 RNA showed binding of WTAPP1 RNA to whole CNBP or truncated CNBP. Schematic of the domain structures of CNBP protein is shown in E and Western blot analysis of FLAG-tagged full-length CNBP or truncated CNBP in 293T cells is shown in F. G, IGV snapshots showed the CNBP CLIP-seq and miCLIP-seq reads distribution on WTAPP1 RNA. Purple tracks are unique tag coverage of CNBP CLIP-seq and blue tracks are unique tag coverage of miCLIP-seq, respectively. Filled blue circle denotes miCLIP-called m6A site. H, The levels of CNBP bound to WTAPP1 RNA in PDAC cells with or without METTL3 knockdown. I, The levels of CNBP bound to WTAPP1 RNA in PDAC cells with METTL3 knockdown but restored by transfection with WT METTL3 or its mutant. J, Binding levels of CNBP with WTAPP1 RNA in PDAC cells cotransfected with dCas13b-ALKBH5 and NT-gRNA or sgRNAs. K and L, Comparison of WTAPP1 RNA level and its stability in cells with or without CNBP silence. M, Immunoprecipitation (IP) assays show CNBP and HuR interaction in PDAC cells. N, The levels of HuR bound to WTAPP1 RNA in cells with or without CNBP silence. O, The levels of HuR bound to WTAPP1 RNA in cells with dm6ACRISPR system. All data are mean ± SEM from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant of Student t test.