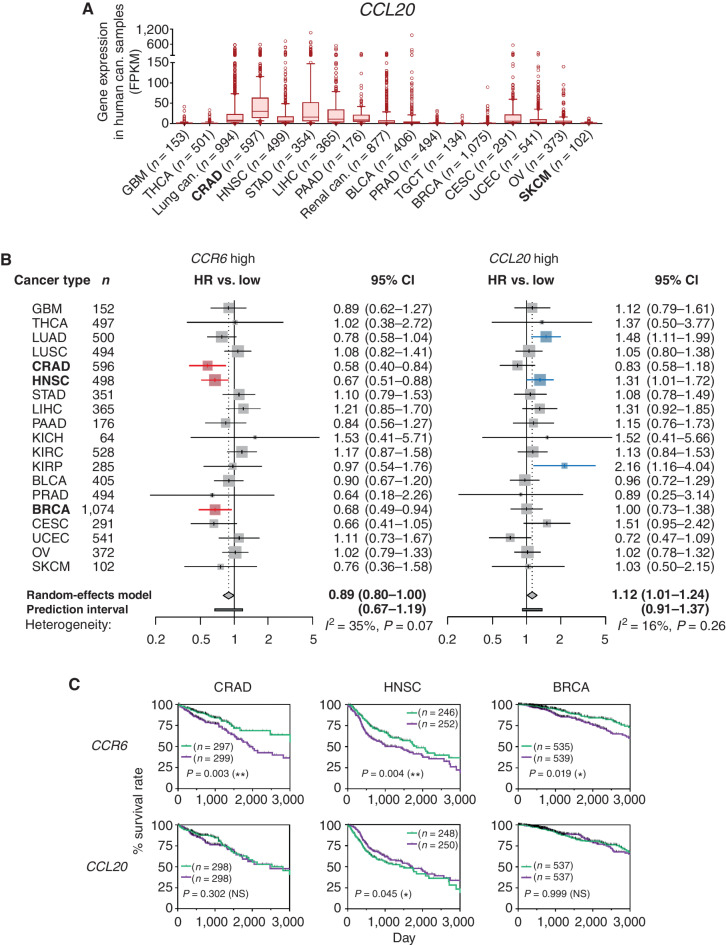
Figure 2.
CCR6 gene expression correlates with favorable prognoses of some human CCL20-producing cancers. A, Fragments per kilobase of exon per million reads mapped (FPKM) of CCL20 gene expression were calculated from the RNA-sequencing results in 20 types of indicated human cancers using The Cancer Genome Atlas (TCGA) database obtained from the Human Protein Atlas. Results are presented as box plots with 10 to 90 percentiles, and outliers are shown. Lung cancer includes LUAD and LUSC, and renal cancer includes KICH, KIRC, and KIRP. B, HRs of death for high CCR6 and CCL20 gene expression in 19 types of human cancers in the TCGA database are presented as forest plots. Results are presented with 95% CIs on a logarithmic scale. Size of the marks indicates the number of samples (patients). The red bars indicate the correlation with prolonged survival, and the blue bars indicate the correlation with brief survival. A random-effects model meta-analysis was applied to pool the effect size, and the pooled effect size with the associated 95% CIs is described in a diamond. The 95% prediction interval of the pooled effect is described in the horizontal bar. Between-cancer-type heterogeneity variance (I2 value) is shown in the panel. C, OS curves for patients with a higher level of CCR6 or CCL20 gene expression in tumor than the median level (green) and patients with lower level of either gene in their tumor than the median level (purple) in CRAD, HNSC, and BRCA are presented. Survival curves are presented within day 3,000, and points censored are shown as a black bar. Results were analyzed with Kaplan–Meier survival analysis by log-rank methods, and P values are shown in the panel. NS, not significant. *, P < 0.05; **, P < 0.01. Number (n) of tumors (A) or patients (B and C) in every type of cancer is indicated. Cancer-type abbreviations are defined in Supplementary Table S1. Can., cancer. Bold font in A and B is used to highlight the most significant findings.