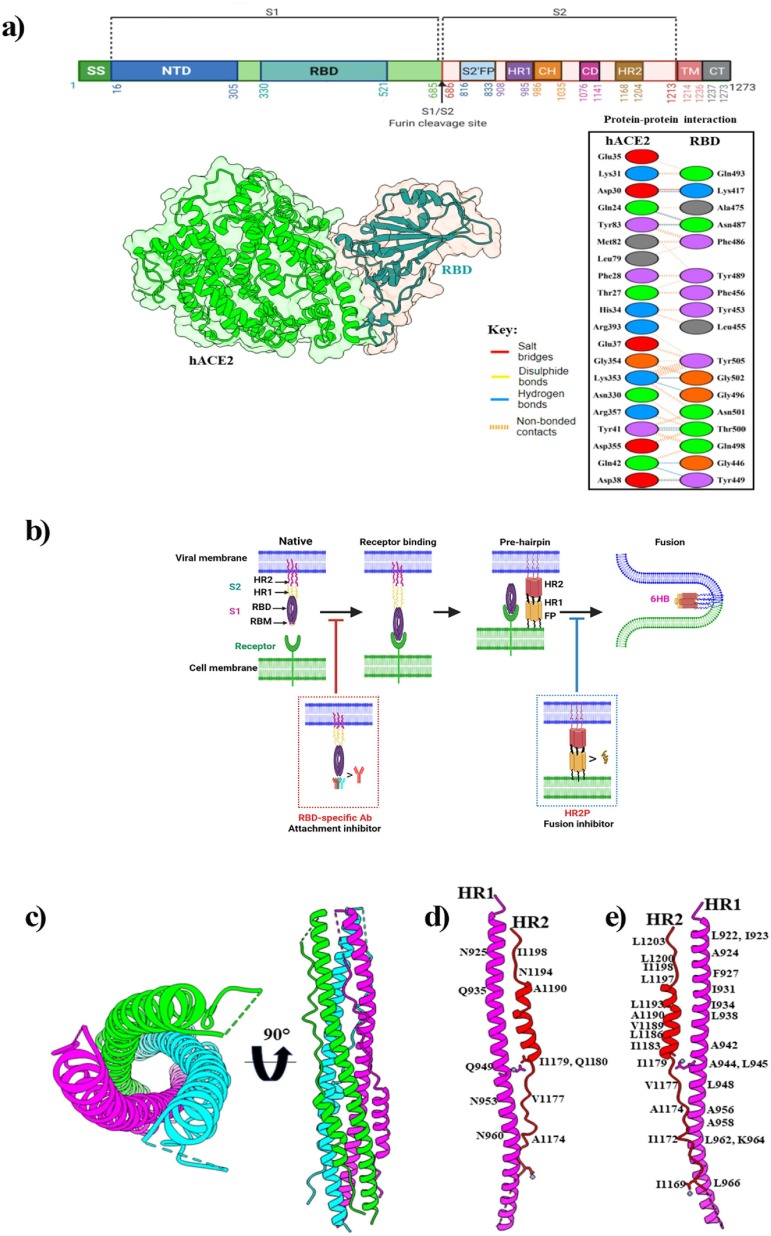
Fig. 3.
(a) Schematic representation of SARS-CoV-2 spike protein primary structure and hACE2-RBD complex. Here, the top panel shows different colors domains. SS, single sequence; NTD, N-terminal domain; RBD, receptor-binding domain; S1, subdomain 1; S2, subdomain 2; S1/S2, S1/S2 protease cleavage site; S2′, S2′ protease cleavage site; FP, fusion peptide; HR1, heptad repeat 1; CH, central helix; CD, connector domain; HR2, heptad repeat 2; TM, transmembrane domain; CT, cytoplasmic tail. Arrows indicate the protease cleavage site. The lower-left panel shows the cartoon and surface representations of the overall structure of the SARS-CoV-2 RBD bound to hACE2 (PDB: 6M0J) and protein–protein interactions are presented in the lower-right panel. (b) The mechanism of human ACE2 and SARS-CoV-2 S protein-mediated virus attachment and fusion. In the native state, the S2 subunit is encapsulated by the S1 subunit. Several conformational changes occur in the S2 subunit after viral RBD engagement with the receptor. The HR1-trimer core structure is formed from three HR1 molecules, and three HR2 molecules bind to the HR1-trimer to form 6-HB, which mediates membrane fusion. A neutralizing antibody that targets RBD blocks viral infection by blocking RBD’s interaction with cellular receptors. The membrane fusion process is inhibited by the fusion inhibitor that blocks 6-HB formation. (c) Six-helix bundle fusion core is comprised of three HR2-helices packed in the HR1 side grooves (PDB: 6M1V). Structures in the top view (left panel) and side view (right panel) are presented respectively. Here, three HR1/HR2 chains are colored light green, magenta, and cyan. (d) The detailed interactions between HR1 and HR2, and residues involved in the H-bond interactions are labelled. (e) Residues involved in the hydrophobic interactions are labeled.