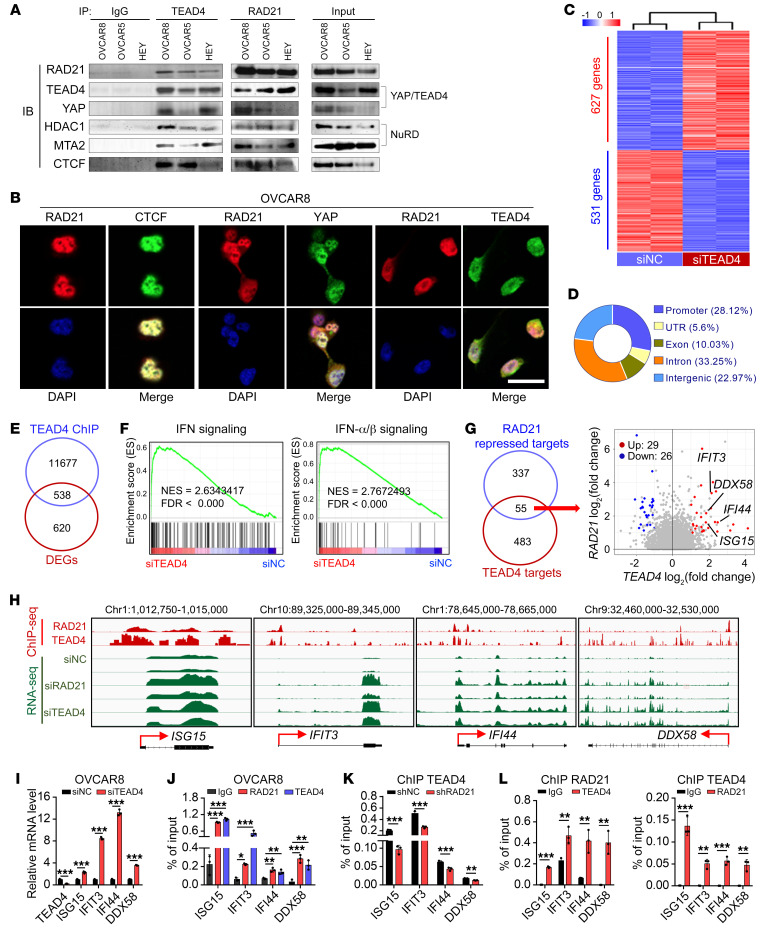
Figure 3. RAD21 directly interacts with YAP/TEAD4 transcriptional corepressor complex to coordinately suppress ISGs.
(A) Coimmunoprecipitation by anti-IgG, anti-TEAD4, and anti-RAD21 antibodies followed by immunoblotting (IB) with antibodies against the indicated proteins using cell extracts from OVCAR8, OVCAR5, and HEY cells. (B) Colocalization of RAD21 with CTCF (left, positive control), YAP (middle), and TEAD4 (right) was visualized by immunofluorescence. Scale bar: 20 μm. (C) Heatmap for DEGs (P < 0.01, |log2foldchange| > 1) in TEAD4-KD and control OVCAR8 cells (duplicates). (D) Genomic distribution of TEAD4 peaks in OVCAR8 cells. (E) Venn diagram showing the overlaps of DEGs and TEAD4 direct targets obtained from TEAD4 ChIP-Seq results. (F) GSEA analysis showing that the IFN Signaling and IFN-α/β Signaling pathways were enriched in TEAD4-KD cells compared with control cells. (G) Venn diagram (left) and scatterplot (right) showing the precise targets repressed by RAD21 and TEAD4. (H) Genome browser tracks of RAD21 and TEAD4 ChIP-Seq and RNA-Seq at genomic loci of ISG15, IFIT3, IFI44, and DDX58. (I) qRT-PCR validation of representative ISGs in TAED4-KD and control OVCAR8 cells. (J) ChIP-qPCR analysis of RAD21 and TEAD4 occupancy at genomic loci of ISGs ISG15, IFIT3, IFI44, and DDX58 in OVCAR8 cells. Data are shown as mean ± SD (n = 3, 1-way ANOVA). (K) ChIP-qPCR analysis of TEAD4 occupancy at genomic loci of ISGs ISG15, IFIT3, IFI44, and DDX58 in RAD21-KD and control OVCAR8 cells mediated by shRNA. (L) Re-ChIP analysis showing the concurrent presence of both RAD21 and TEAD4 at genomic loci of ISGs ISG15, IFIT3, IFI44, and DDX58. Data in I, K, and L are shown as mean ± SD (n = 3, 2-tailed t test). *P < 0.05; **P < 0.01; ***P < 0.001.