Abstract
Hundreds of leucine-rich repeat receptor kinases (LRR-RKs) have evolved to control diverse processes of growth, development, and immunity in plants; the mechanisms that link LRR-RKs to distinct cellular responses are not understood. Here we show that two LRR-RKs, the brassinosteroid hormone receptor BRI1 (BRASSINOSTEROID INSENSITIVE 1) and the flagellin receptor FLS2 (FLAGELLIN SENSING 2), regulate downstream glycogen synthase kinase 3 (GSK3) and mitogen-activated protein (MAP) kinases, respectively, through phosphocoding of the BRI1-SUPPRESSOR1 (BSU1) phosphatase. BSU1 was previously identified as a component that inactivates GSK3s in the BRI1 pathway. We found surprisingly that loss of the BSU1 family phosphatases activates effector-triggered immunity (ETI) and impairs flagellin-triggered MAP kinase activation and immunity. The flagellin-activated BOTRYTIS-INDUCED KINASE 1 (BIK1) phosphorylates BSU1 at serine-251. Mutation of serine-251 reduces BSU1’s ability to mediate flagellin-induced MAP kinase activation and immunity, but not its abilities to suppress ETI and interact with GSK3, which is enhanced through the phosphorylation of BSU1 at serine-764 upon brassinosteroid signaling. These results demonstrate that BSU1 plays an essential role in immunity and transduces brassinosteroid-BRI1 and flagellin-FLS2 signals using different phosphorylation sites. Our study illustrates that phosphocoding in shared downstream components provides signaling specificities for diverse plant receptor kinases.
The gene family of leucine-rich repeat receptor kinases (LRR-RK) has expanded dramatically in higher plants, with over 200 and 400 members in Arabidopsis and rice, respectively1. Among the Arabidopsis LRR-RKs, the brassinosteroid (BR) receptor BRI1 and the flagellin receptor FLS2 have distinct functions but are mechanistically related. Activation of BRI1 by BR and activation of FLS2 by the pathogen-associated molecular pattern (PAMP) flagellin involve ligand-induced association with the common co-receptor BRI1-ASSOCIATED KINASE1 (BAK1)2–4. Signal transduction downstream of BRI1 is mediated by a well-characterized cascade of phospho-relay events, which include BRI1-mediated phosphorylation of the BR-SIGNALING KINASE (BSK) and CONSTITUTIVE DIFFERENTIAL GROWTH 1 (CDG1) family of receptor-like cytoplasmic kinases (RLCKs)5, CDG1-mediated phosphorylation of serine-764 (S764) of the BRI1-SUPPRESSOR1 (BSU1) phosphatase6, and BSU1-mediated dephosphorylation of the GSK3 kinase BRASSINOSTEROID-INSENSITIVE 2 (BIN2)7, leading to SERINE/THREONINE PROTEIN PHOSPHATASE 2A (PP2A)-mediated dephosphorylation of the BRASSINAZOLE-RESISTANT 1 (BZR1) family of transcription factors8,9. Signaling pathways downstream of FLS2 also involve RLCKs such as BIK1 and PBS1-LIKE (PBL) kinases10,11, which activate the MAP kinases through mechanisms that are not fully understood12. In addition to sharing the co-receptor kinase BAK1, BRI1 and FLS2 have been shown to also share their substrates BSK1 and BIK1 kinases13,14. The molecular mechanisms that ensure signaling specificity of these shared downstream components remain unknown.
Plant immune responses are mediated by two systems: pattern-triggered immunity (PTI) and effector-triggered immunity (ETI). PTI is activated by pattern recognition receptors (PRRs) that recognize PAMPs at the cell surface. Pathogens deliver effector proteins into plant cells to attenuate plant PTI. Plants then evolved intracellular receptors to recognize the perturbations of PTI pathways by pathogen effectors15. Recent studies showed that PTI and ETI activate overlapping downstream defense outputs and mutually potentiate each other16–18. Genetic perturbations of some PTI components, such as FLS2’s co-receptor BAK1 and substrate BIK1, activate ETI and increase resistance to virulent bacterial pathogens19,20.
The Arabidopsis BSU1 family of phosphatases includes 4 members named BSU1 and BSU1-LIKE 1 (BSL1) to BSL3 (together named BSLs)21,22. In addition to mediating BR signaling, The BSU1 family also plays an essential role in stomatal development22. BSLs promote stomatal asymmetric cell division by establishing kinase-based signalling asymmetry in the two daughter cells34. The partial loss-of-function quadruple mutant (bsu1;bsl1;amiBSL2/3, named bsu-q) displays pleiotropic defects and the bsl1;bsl2,bsl3 triple mutant is embryo lethal, which suggest additional roles beyond BR signaling and stomata development22,23.
Results
BSU1 family plays an essential role in immunity
To fully understand the functions of the BSU1 family phosphatases, we performed RNA-seq analysis of bsu-q. The results show not only decreased expression of growth-related genes as expected but also increased expression of genes related to immunity in bsu-q (Supplementary Table 1 and 2; Extended Data Fig. 1).
To understand the function of BSLs in immunity, we analyzed how bsu-q affects the expression levels of genes that are activated during ETI16 and PTI (treatment with flagellin peptide flg22, Supplementary Table 3). Consistent with the GO analysis, more ETI- and flg22-induced genes were increased (178) than decreased (56) in bsu-q. Compared to wild type, bsu-q increased the expression levels (>2x) of a large portion (82 of 823) of the genes activated by both ETI and flg22, and smaller but significant portions of genes activated by only ETI (56 of 1374) or only flg22 (40 of 547) (Fig. 1a). Many flg22-insensitive but ETI-activated genes were dramatically increased in bsu-q (Fig. 1b). In contrast, many ETI-independent but flg22-induced genes, including 11 of the 53 genes induced over 5-fold by flg22 in wild-type plants, showed no flg22 response in bsu-q (Fig. 1c; Supplementary Table 4). The bsu-q mutant showed a reduced flg22 responsiveness at the transcriptomic level (Supplementary Table 3; Extended data Fig. 2a, b). Quantitative PCR analysis confirmed the RNA-seq results for several selected genes that showed elevated expression levels (FLG22-INDUCED RECEPTOR-LIKE KINASE 1 (FRK1), At2g17740, and CYSTEINE-RICH RLK 13 (CRK13)) or insensitivity to flg22 (ROF2) in bsu-q (Extended data Fig. 2c). These results suggest that the bsu-q mutant has a constitutive activation of ETI response and a defect in flagellin signaling. Similar phenotypes have been observed in mutants lacking components of PAMP-signaling, such as the bak1 bkk1 mutant which lacks the co-receptor for FLS2 and BRI1. Indeed, a comparison of transcriptomes affected in bak1 bkk119 with those affected in bsu-q revealed that the mutants share similar gene expression patterns (Fig. 1d). Of the 509 genes upregulated in bsu-q, 240 (47%) were also upregulated in bak1 bkk1, and these include 132 (55%) ETI- and/or flg22-induced genes (Fig. 1d). The transcriptomic analyses support a possibility that BSU1 family phosphatases play dual roles in immunity and BR signaling, similar to that observed for the upstream signaling components BAK1, BIK1, and BSK113,14,19.
Fig. 1. The bsu-q mutant shows enhanced ETI and defective flagellin signaling.
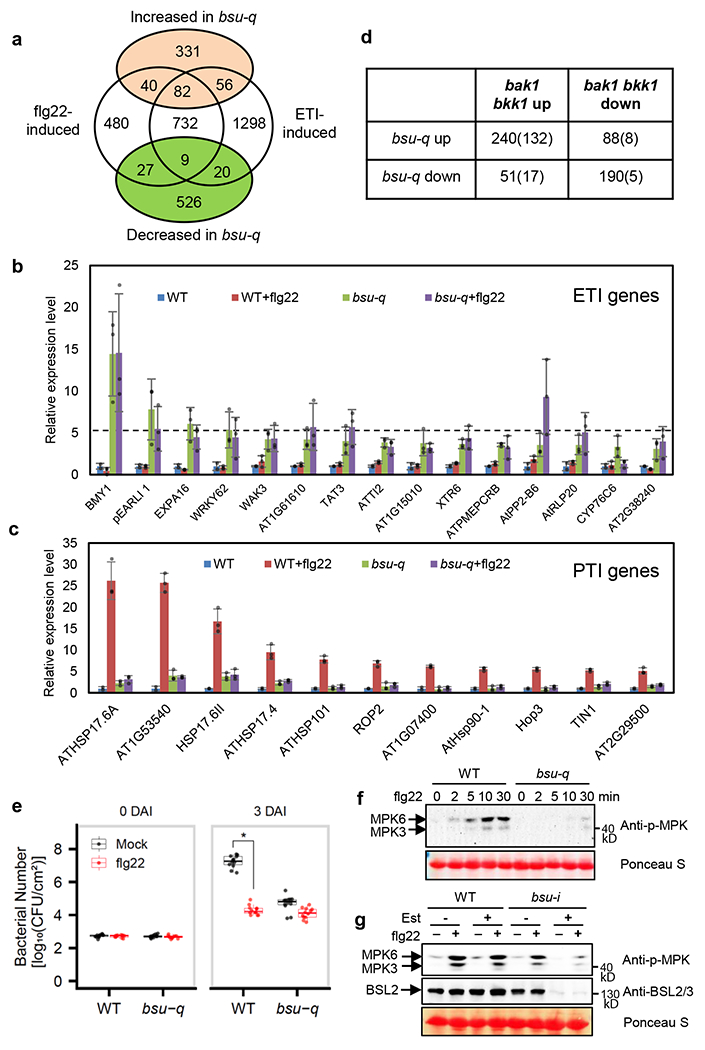
(a) The Venn diagram shows the overlaps between the genes up- or down-regulated in bsu-q relative to wild type and the genes activated by flg22 treatment or ETI. (b-c) Relative expression levels normalized to wild type (WT) of (b) ETI genes that did not respond to flg22 in wild type but increased in bsu-q, and (c) 11 genes, among the 53 ETI-independent genes that were induced over 5-fold by flg22 in WT but showed no significant response to flg22 (FC <2) in bsu-q. The graph shows mean ± SEM values from the RNA-seq data in Supplementary Table 4. (d) Overlap between differentially expressed genes of bsu-q and bak1 bkk1. Numbers in parenthesis are ETI/flg22-responsive genes. (e) Quantitation of Pseudomonas syringae pathovar tomato strain DC3000 in WT and bsu-q plants treated with mock or flg22. Each replicate includes four leaf discs and 12 biological repeats (n=12) were used, and the experiment was repeated three times with similar results. The box plot shows the minimum, first quartile, median, third quartile and maximum of the 12 displayed data points for each sample. Asterisk indicates statistically significant difference by two-tail t-test (p=7.115e-16). (f) The bsu-q mutant is defective in flg22-induced MPK3/6 phosphorylation. Leaf tissues of wild type (WT) and bsu-q were treated with 100 nM flg22 for the indicated time, and MAP kinase activation was analyzed by immunoblot using anti-phospho-p44/42 MAPK antibody. Ponceau S staining of membrane shows equal protein loading. The experiment was repeated three times with similar results. (g) WT and bsu-i seedlings were grown on medium containing 0 or 20 μM estradiol for nine days, and then treated with 100 nM flg22 for 10 min. Immunoblots were probed with anti-phospho-p44/42 MAPK and anti-BSL2/3 antibodies. Ponceau S staining of membrane shows equal protein loading. The experiment was repeated twice with similar results.
To test whether the altered defense gene expression affects immune response in bsu-q, we analyzed bacterial growth after inoculation of wild type and bsu-q mutant with Pseudomonas syringae pathovar tomato (Pst) strain DC3000. The results show that bsu-q is more resistant than wild type to Pst infection (Fig. 1e). Pretreatment of seedlings with flg22 inhibited bacterial growth in the wild-type plants but did not further enhance disease resistance in the bsu-q mutant (Fig. 1e). The priming of defense responses in untreated bsu-q plants and compromised flg22-triggered responses are consistent with the altered expression of ETI and PTI genes. These phenotypes are similar to those of other mutants with loss of PAMP signaling components, including the bik120 and bak1 bkk1 double mutant19. The bak1 bkk1 double mutant exhibits impaired PTI responses such as flagellin-induced MAP kinase activation and reactive oxygen response (ROS), as well as enhanced ETI and accumulation of the defense hormone salicylic acid (SA)19. Similarly, we observed a weaker flg22-induced ROS response in the bsu-q mutant and an elevated level of SA in the inducible quadruple mutant bsu-i plants (bsu1;bsl1;bsl3 triple mutant transformed with a construct for estradiol-inducible expression of an artificial microRNA targeting BSL2. See Methods) (Extended Data Fig. 3). These defense-related phenotypes similar to the bak1 bkk1 and bik1 mutants further support the hypothesis that BSU1 family members have dual functions, similar to BAK1 and BIK1, as components of PTI signaling pathways and guardees of resistance proteins operating in ETI signaling pathways. In line with this hypothesis, a recent study identified the BSU1 family members as targets of pathogen effector proteins24.
BSU1 mediates flagellin signaling as a BIK1 kinase substrate
To determine whether the BSU1 family members play a role in FLS2 signaling, we analyzed flg22-induced activation of MAP kinases in mutants lacking various BSU1/BSLs. Flg22 treatment induced similar MAP kinase phosphorylation in wild type and bsl2;bsl3 double mutant (Extended Data Fig. 4a), but induced much weaker MAP kinase phosphorylation in the bsu-q mutant (Fig. 1f) and in the bsu-i plants (Fig. 1g), indicating that BSU1/BSLs are redundantly required for flg22-induced MAP kinase activation. In addition, MAP kinase phosphorylation caused by another bacterial elicitor, elf1825, was reduced in the estradiol-treated bsu-i mutant (Extended Data Fig. 4c), suggesting a more general role for BSU1/BSLs in PTI signaling.
The mode of function of BSU1 in the BR signaling pathway suggests that BSU1 may transduce the flagellin signal from BIK1 to downstream MAP kinases. We thus performed yeast-two-hybrid assays to test if BSU1/BSLs interact with BIK1 and MEKK1, a MAP kinase kinase kinase (MAPKKK) involved in flagellin-induced MAP kinase activation26,27. The results show that BSU1 interacts with both BIK1 and MEKK1 (Fig. 2a,b) and that BSU1/BSLs interact with multiple BIK1/PBL proteins with various specificities and affinities (Extended Data Fig. 5), suggesting that different BSU1 and BIK1/PBL family members may act redundantly in parallel pathways. Consistent with the yeast two-hybrid assay results, in vitro pull-down assays showed that BIK1 interacts with BSU1 but not the BSLs (Fig. 2c), and BSU1 also interacts with MEKK1 in vitro (Fig. 2d). Moreover, co-immunoprecipitation assays confirmed the interaction between BIK1-Myc and BSU1-YFP in Nicotiana benthamiana and transgenic Arabidopsis plants (Fig. 2e), as well as between BSU1-Myc and MEKK1-YFP (Fig. 2f). These results suggest a potential role for BSU1 in transducing the signal from BIK1 to MEKK1.
Fig. 2. BSU1 interacts with BIK1 and MEKK1.
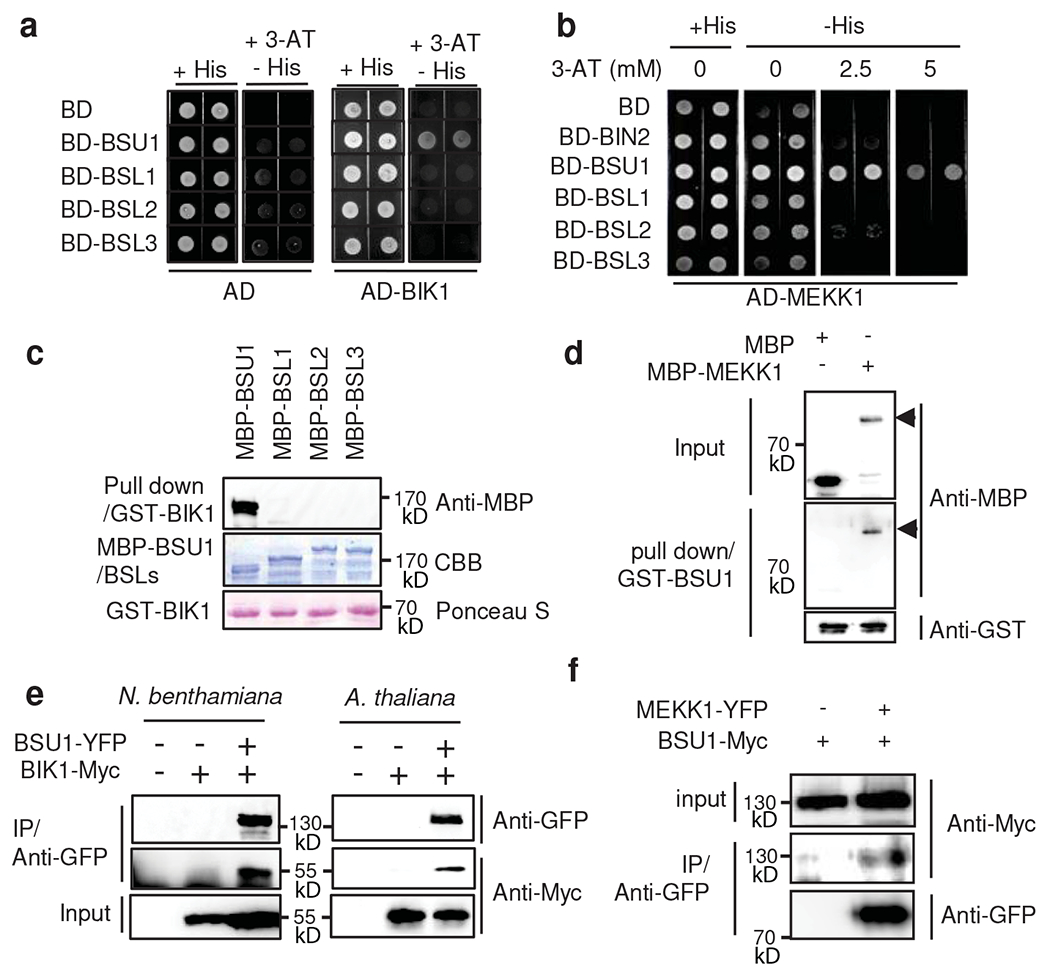
(a) Yeast-two-hybrid assays of interaction between BIK1 and BSU1/BSLs. Yeast cells co-expressing AD-BIK1 and BD-BSU1/BSLs were grown on synthetic dropout medium containing 2.5 mM 3-amino-1, 2, 4-triazole (3-AT). (b) Yeast-two-hybrid assays of interaction between MEKK1 and BSU1/BSLs. (c) In vitro pull-down of BSU1/BSLs (as MBP fusions) by GST-BIK1, analyzed by anti-MBP immunoblotting. Input proteins are shown by Coomassie brilliant blue (CBB) of the SDS-PAGE gels and Ponceau S staining of membranes. The experiment was repeated twice with similar results. (d) In vitro pull-down of MBP-MEKK1 by GST-BSU1, analyzed by immunoblotting using anti-MBP and anti-GST antibodies. Arrow heads indicate MBP-MEKK1. The experiment was repeated twice with similar results. (e) Co-immunoprecipitation assay of BSU1-YFP with BIK1-Myc protein expressed in N. benthamiana leaves and Arabidopsis transgenic plants. The experiment was repeated twice in Arabidopsis with similar results, (f) Co-immunoprecipitation assay of BSU1-Myc with MEKK1-YFP protein expressed in N. benthamiana leaves. The experiment was repeated twice with similar results.
The interaction with BIK1 suggests that BSU1 might be phosphorylated upon flagellin signaling. To test this possibility, we performed metabolic Stable Isotope Labeling In Arabidopsis followed by Immuno-Precipitation and Mass Spectrometry (SILIA IP-MS) analysis of BSU1-YFP in transgenic Arabidopsis. We found that flg22 induced phosphorylation of the serine-251 (S251) residue of BSU1 (Fig. 3a,b). Targeted quantification using parallel reaction monitoring (PRM) mass spectrometry further showed that phosphorylation of BSU1 at S251 was increased over 4 fold after flg22 treatment in both replicate experiments (Fig. 3c,d and Extended Data Fig. 6a,b). The S251 residue of BSU1 is located within the N-terminal Kelch-repeat domain and is conserved in BSL homologs, and paralogs in other plant species (Fig. 3e, Extended Data Fig. 7).
Fig. 3. Flagellin induces phosphorylation of BSU1 at S251.
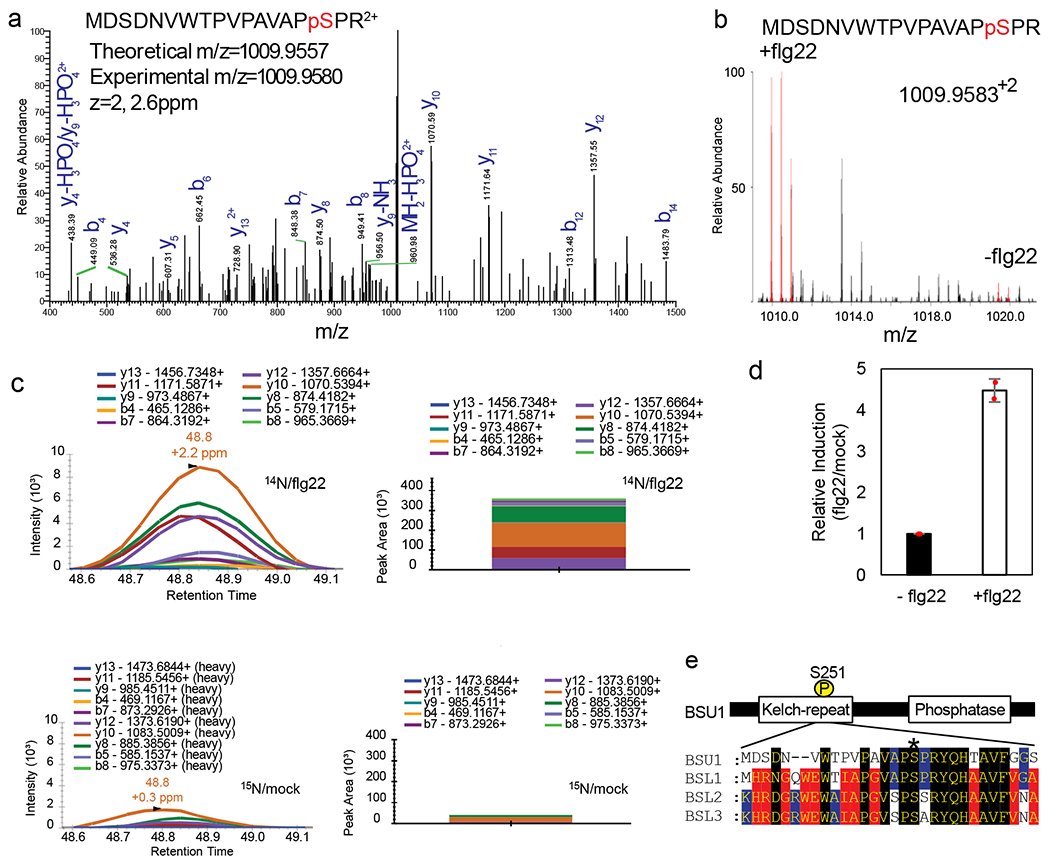
(a) MS/MS spectrum of the peptide containing phosphorylated S251 of BSU1. (b) MS1 spectrum of a SILIA IP-MS experiment shows flg22-induced phosphorylation of BSU1 S251. The peaks of 14N-labeled (+flg22) and 15N-labeled (−flg22) phospho-S251-containing peptides are shown in red. (c) PRM quantitation of the phospho-S251 peptide after SILIA and immunoprecipitation of BSU1-YFP from mock-treated 15N-labeled and flg22-treated 14N-labeled seedlings. Graphs display chromatograms of fragment ions extracted from the 14N- and 15N-labeled phospho-peptides. Peak areas of fragment ions are extracted using <5ppm mass window and integrated across the elution profile. (d) Quantification of relative induction (flg22/mock). The graph shows mean ± SEM from two reciprocal SILIA IP-MS experiments (n=2). (e) Alignment of sequences of BSU1 family members around the BSU1 S251 residue (asterisk).
We next tested whether BIK1 phosphorylates BSU1 using in vitro kinase assays. As shown in Figure 4a, GST-BIK1 phosphorylated MBP-BSU1 specifically but not the BSLs, consistent with the yeast interaction results (Fig. 2a). GST-BIK1 phosphorylated the N-terminal Kelch domain more strongly than the C-terminal phosphatase domain (Fig. 4b). The MBP-BSU1 protein containing mutation of S251 to alanine (S251A) was less phosphorylated than wild-type BSU1 by GST-BIK1 (Fig. 4c). In contrast, the alanine substitution of the CDG1 phosphorylation site (S764A) had no obvious effect on BIK1 phosphorylation of BSU1 (Fig. 4c). As reported previously28, flg22 treatment induced phosphorylation of BIK1, causing a slower mobility in SDS-PAGE gel and enhanced autophosphorylation (Fig. 4d). BIK1-HA immunoprecipitated from the flg22-treated plants showed stronger phosphorylation of and interaction with MBP-BSU1 than that from untreated plants (Fig. 4d,e). The phosphorylation of BSU1 by BIK1-HA was also decreased by the S251A mutation (Fig. 4f). These results demonstrate that BIK1 phosphorylates the S251 residue of BSU1 in response to flagellin-FLS2 signaling.
Fig. 4. BIK1 phosphorylates BSU1 at S251.
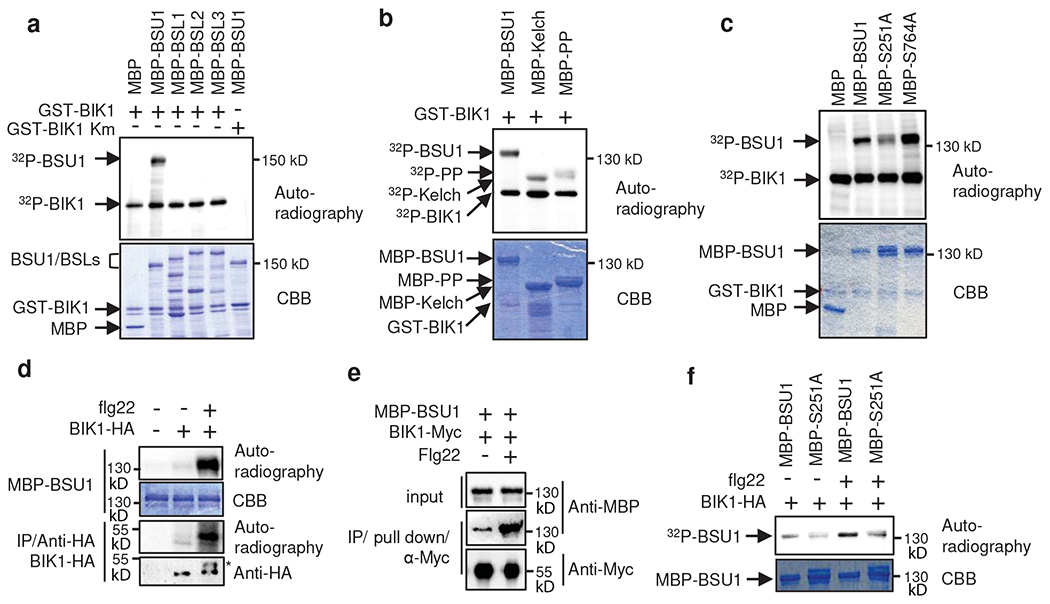
(a) In vitro kinase assay of BIK1 phosphorylation of the indicated MBP and MBP fusion proteins. Phosphorylation and proteins are shown by autoradiography and gel staining (CBB), respectively. A kinase inactive mutant (Km) BIK1 was used as a negative control. The experiment was repeated three times with similar results. (b) In vitro kinase assay of BIK1 phosphorylation of BSU1 N-terminal (Kelch) and C-terminal (PP) domains. The experiment was repeated twice with similar results. (c) In vitro kinase assay of GST-BIK1 phosphorylation of wild-type and mutant (S251A and S764A) MBP-BSU1. The experiment was repeated three times with similar results.
(d) In vitro kinase assay of flg22-induced BIK1 phosphorylation of BSU1. BIK1-HA was immunoprecipitated using anti-HA antibody from thirteen-day old BIK1::BIK1-HA seedlings treated with 1 μM flg22 or mock for 10 min and then incubated with MBP-BSU1 and 32P-γ-ATP. BIK1-HA proteins were analyzed by immunoblotting using anti-HA antibody. Asterisk indicates phosphorylated BIK1. The experiment was repeated three times with similar results. (e) Flg22-induced BIK1-BSU1 interaction. BIK1-Myc proteins were immuno-purified by anti-Myc antibody from plants treated with 100 nM flg22 or mock for 30 min and were used to pulldown MBP-BSU1 in vitro. The experiment was repeated twice with similar results. (f) In vitro kinase assay of flg22-induced BIK1 phosphorylation of wild-type and mutant (S251A) BSU1 using immuno-purified BIK1-HA. BIK1-HA was immunoprecipitated using anti-HA antibody from twelve-day-old BIK1::BIK1-HA seedlings treated with 1 μM flg22 or mock for 10 min, and then incubated with MBP-BSU1 or MBP-BSU1-S251A (MBP-S251A) and 32P-γ-ATP. The experiment was repeated twice with similar results.
Phosphorylation of BSU1-S215 mediates MAP kinase activation
To determine whether phosphorylation of S251 in BSU1 plays a role in flagellin-induced MAP kinase activation, we generated transgenic bsu-i plants expressing BSU1-YFP or BSU1-S251A-YFP. Two independent transgenic lines expressing wild-type BSU1 in bsu-i increased MAP kinase activation by more than 1.7-fold, whereas expression of BSU1-S251A only increased MAP kinase by about 1.3-fold (Fig 5a; Extended Data Fig. 8a).
Fig. 5. Phosphorylation of BSU1 S251 is required for MAPK activation and immunity but not for BR signaling.
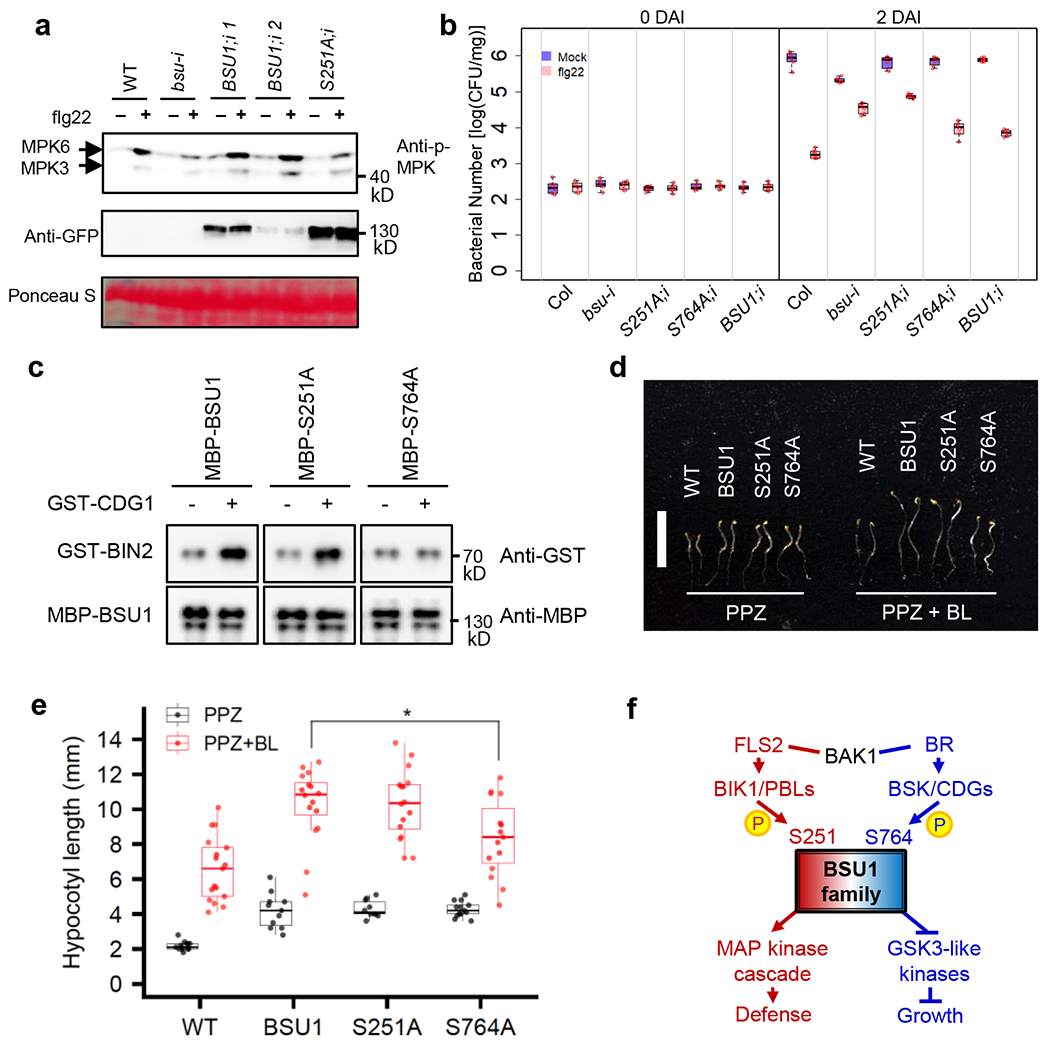
(a) Flg22-induced MPK3/6 phosphorylation in 35S::BSU1-YFP;bsu-i (BSU1;i) and 35S::BSU1-S251A-YFP;bsu-i (S251A;i) plants. Eight-day old seedlings grown on 20 μM estradiol were treated with 10 nM flg22 for 15 min. Immunoblot was performed using anti-phospho-p44/42 MAPK and anti-GFP antibodies. Ponceau S staining image shows equal loading. The experiment was repeated three times with similar results. (b) Flg22-induced bacterial growth inhibition in seedlings of the indicated genotypes at 0 and 2 days after inoculation (DAI). Each replicate includes 20 mg or more seedlings and 6 biological replicates. The box plot shows the minimum, first quartile, median, third quartile and maximum of the 6 displayed data points for each sample. (c) CDG1-induced BIN2 interaction with MBP-BSU1, MBP-BSU1-S251A and MBP-BSU1-S764A. Wild type and mutant MBP-BSU1 proteins pre-incubated with GST(−) or GST-CDG1(+) were used to pull down GST-BIN2. The experiment was repeated three times with similar results. (d, e) Phenotype (d) and hypocotyl length (e) of 5-day-old etiolated WT, 35S::BSU1-YFP (BSU1), 35S::BSU1-S251A-YFP (S251A) and 35S::BSU1-S764A-YFP (S764A) seedlings grown on media containing PPZ or PPZ plus BL. The box plot shows the minimum, first quartile, median, third quartile and maximum of the displayed data points for each sample. (d) Bar = 10 mm. (e) The average hypocotyl length ± SEM is shown. Asterisk denotes significant difference from 35S::BSU1-YFP by two-tailed t-test (p-value =0.01512, n = 16 for BSU1 and n = 15 for S764A). Experiments were performed 3 times and similar results were obtained. (f) Schematic diagram of the BSU1 phosphocodes in BRI1 and FLS2 signaling pathways.
We then tested whether the S251A mutation affects BSU1’s ability to mediate flagellin-induced immunity against pathogen infection. We treated seedlings with mock or flg22 peptide, inoculated them with Pst DC3000, and then quantified the number of bacteria in the plant tissue at zero and two days after the inoculation. Flg22 treatment decreased the growth of bacteria by about 464-fold in wild type plants. The mock-treated bsu-i plants contained a reduced number of bacteria compared to mock-treated wild type, whereas flg22 treatment caused only about 6-fold reduction of the bacteria growth in bsu-i, which was similar to bsu-q. These phenotypes are consistent with activation of ETI signaling and a compromise in PTI signaling due to the loss of BSU/BSL family members. The bsu-i plants expressing either wild-type BSU1, BSU1-S251A, or BSU1-S764A showed similar pathogen growth as wild type after mock treatment, suggesting that both the wild type and mutant BSU1 proteins rescued the ETI phenotype. Upon flg22 treatment, the BSU1-251A/bsu-i plants showed only about 9-fold reduction of pathogen growth, whereas the BSU1/bsu-i and BSU1-S764A/bsu-i plants showed 110-fold and 72-fold pathogen reduction compared to mock treatment (Fig. 5b). These data indicate that both wild type and mutant BSU1 proteins can suppress the ETI response, whereas BSU1’s function in PTI response is impaired by the S251A mutation but not the S764A mutation. These results further support the conclusions that the presence of BSU1/BSL proteins is required to control ETI, and the BIK1 phosphorylation of BSU1 at S251 contributes to flg22 induction of MAP kinase activation and immunity, whereas phosphorylation of S764 does not play a major role in immune signaling.
Previous studies have shown that BR-BRI1 signaling involves CDG1-mediated phosphorylation of BSU1 S764, which enhances interaction with BIN2, a GSK3-like kinase6. To test whether phosphorylation at S251 and S764 specifies downstream targets of BSU1 signaling, we compared the effects of mutations of these residues on BSU1 interaction with BIN2. Consistent with the previous report, phosphorylation by CDG1 increased the interaction of wild-type BSU1 with BIN2 but not that of the BSU1-S764A mutant protein (Fig. 5c). The BSU1-S251A mutant, similar to wild-type BSU1, showed CDG1-induced interaction with BIN2 (Fig. 5c, Extended Data Fig. 8b). Consistent with the normal interaction with BIN2, seedlings expressing BSU1-S251A showed similar BR sensitivity in hypocotyl elongation compared to plants expressing wild type BSU1, whereas plants expressing BSU1-S764A showed reduced sensitivity to BR (Fig. 5d,e, Extended Data Fig. 9). These results show that BSU1’s functions in the flagellin and BR pathways require phosphorylation of distinct residues, S251 and S764, respectively.
Discussion
Taken together our results show that BSU1 and BSLs redundantly play key roles in plant immunity, in addition to their known functions in BR signaling and stomata development. The presence of a BSU1/BSL protein is required to suppress immune responses and the loss of all BSU1 family members triggers an ETI-like response, whereas flagellin-FLS2 signaling induces BIK1 phosphorylation of BSU1 at S251 which leads to activation of the MAP kinases and immune responses. The mechanism by which phospho-S251 leads to activation of the MAP kinases remains to be investigated. Previous studies have shown that several MEKKs are inhibited by phosphorylation of their N-terminal auto-regulatory domain, suggesting that activation of MEKKs involves dephosphorylation23,29,30. Our observations of direct interaction between BSU1 and MEKK1 support a model that BSU1 activates MEKK1 by dephosphorylating its auto-regulatory domain.
BR-BRI1 signaling induces CDG1/BSKs-mediated phosphorylation of BSU1 at S764, which mediates interaction with and dephosphorylation of GSK3-like kinases, leading to growth responses14. As such, the signaling specificities for BSU1 in the BRI1 and FLS2 pathways are determined by phosphorylation of different residues, namely phosphocodes (Fig. 5f). Considering the interactions between additional members of the BSU1 and BIK1/PBL families (Extended Data Fig. 5) and the conservation of S251 among BSL family members in Arabidopsis and other plants (Extended Data Fig. 7), the mechanism demonstrated here for Arabidopsis BSU1 is likely used by its paralogs and homologs in parallel/redundant immune pathways. We propose that phosphocoding, namely distinct signal transduction through specific phosphorylation sites, contributes to the diversification of signaling specificity of the large numbers of receptor kinases in plants. In light of the recent finding of spatial recruitment of BSLs in asymmetric cell division31, we propose that phosphocoding and spatial recruitment together provide the signaling specificity that allows the BSU1 family of phosphatases to act in many receptor kinase pathways controlling diverse processes of growth, development, and immunity.
METHODS
Plant material and growth condition
All Arabidopsis thaliana plants used in this study are in Columbia ecotype background. The bsu1-1 (SALK_030721)7, bsl1-1 (SALK_051383)7, bsl2-3 (SALK_055335)22, bsl3-2 (SALK_072437)22, bsu-q7, 35S::BSU1-YFP7, 35S::BSU1-Myc7, BSU1pro::BSU1-YFP32, BIK1::BIK1-HA11, and MEKK1-FLAG27 lines were previously described. The bsl2;bsl3 and bsu1;bsl1;bsl3 mutants were generated by crossing the single mutants. The estradiol-inducible BSU1 family quadruple mutant (bsu-i) was generated by expressing the artificial microRNA targeting BSL2/3 (same as used in bsu-q)3 from the estradiol inducible promoter of pMDC7 vector33 in the bsu1;bsl1;bsl3 triple mutant background. Arabidopsis seedlings were grown on 0.8% phytoblend agar medium containing 1/2 Murashige-Skoog (MS) basal nutrient and 1% sucrose under continuous light in a growth chamber (Percival) at 22 °C. For estradiol treatment, seeds were planted on the MS medium containing β-estradiol (Sigma E2758). For the RNA-seq experiment, seeds were germinated in the dark for 3 days, seedlings with weak phenotypes (long hypocotyl) were removed from the bsu-q samples, and then the plants with strong dwarf phenotypes were grown under light for 12 days. For hypocotyl elongation assay, seeds were planted on MS media containing 2 μM PPZ (Propiconazole, a BR biosynthetic inhibitor, Martin’s HONOR GUARD PPZ)34 or 2 μM PPZ and 1 nM BL (Cayman, 21594), and then stratified for 2 days. After stratification, the seeds were treated with light for 6 hr and then incubated in the dark for 5 days. For general growth and seed harvesting, plants were grown in soil in a greenhouse with a 16-hr light/8-hr dark cycle at 22–24 °C.
Plasmid constructions
To generate the yeast-two-hybrid, 35S::BIK1-Myc, 35S::MEKK1-YFP and MBP-MEKK1 constructs, the full-length coding sequences (CDS) without stop codon of the genes were amplified by PCR from Arabidopsis cDNA using gene-specific primers described in Supplementary Table 6. Each CDS was cloned into pENTR/SD/D-TOPO vector (Invitrogen) and sub-cloned into Gateway-compatible destination vectors (pCAMBIA1390-4Myc-6His, pEarleyGate10135, gcpMALc2x, pXDGATcy86, and pGADT7) by LR reaction. The plasmids for MBP-BSU1, MBP-BSU1-S764A, MBP-BSU1-Kelch, MBP-BSU1-PP, MBP-BSL1, MBP-BSL2, MBP-BSL3, GST-CDG1, GST-BIK1, GST-BIK1Km, 35S::BSU1-YFP, and 35S::BSU1-S764A-YFP were previously described6,7,10. The BSU1 clone in pENTR/SD/D-TOPO vector6 was used as a template for site-directed mutagenesis following the manufacturer’s protocol (New England Biolab) to generate BSU1-S251A, which was subcloned into the Gateway-compatible pEarleyGate101 vector to generate the 35S::BSU1-S251A-YFP construct.
Transgenic plants
The 35S::BSU1-S251A-YFP and 35S::BSU1-S764A-YFP plasmids were transformed into wildtype Arabidopsis using the Agrobacterium-mediated floral dipping transformation method. Transgenic lines expressing similar levels of the protein were selected for experiments shown in Fig. 5d–e and Extended Data Fig. 9. The estradiol-inducible BSU1 family quadruple mutant (bsu-i) was generated by expressing the artificial microRNA targeting BSL2 (same as used in bsu-q)3 from the estradiol inducible promoter of the pMDC7 vector38 in the bsu1;bsl1;bsl3 triple mutant. The 35S::BSU1-YFP;bsu-i (BSU1;i), 35S::BSU1-S251A-YFP;bsu-i (S251A;i), and 35S::BSU1-S764A-YFP;bsu-i (S764A;i) plants were generated by transforming the bsu-i plants with the corresponding plasmids.
PAMP treatment
The flg22 and elf18 peptides were synthesized by the Protein and Nucleic Acid Facility at Stanford University (http://pan.stanford.edu/). The flg22 or elf18 peptides were dissolved in autoclaved distilled water containing 0.05% Silwet L-77 and applied to seedlings or rosette leaves. For RNA-seq experiments, seedlings on plates were submerged in 1 μM flg22 or mock solution for 1 min. After decanting the solution, the seedlings were then incubated in a growth chamber for 1 hr. For Fig. 1f, rosette leaves of eight-week-old short-day-grown plants were treated with 100 nM flg22. For Fig. 1g, nine-day-old seedlings grown on MS media containing 20 μM estradiol were treated with 100 nM flg22 for 10 min. For Fig. 5a, eight-day-old seedlings grown on MS media containing 20 μM estradiol were treated with 10 nM flg22 for 15 min. For Fig. 4d, f, the BIK1::BIK1-HA seedlings were treated with 1 μM flg22 for 10 min.
RNA-seq analysis
Total RNA was extracted from the wild-type and bsu-q seedlings treated with flg22 or mock solution using the Spectrum Plant Total RNA kit (Sigma). cDNA libraries were constructed using a TruSeq RNA sample preparation kit (Illumina) according to the manufacturer’s instructions. Samples of three replicates were sequenced on an Illumina HiSEQ2000 sequencing machine. Mapping of reads to the reference genome (TAIR10) and calling differentially expressed genes was conducted via the iPlant collaborative Discovery Environment (de.iplantcollaborative.org/de/)36. Expression data was clustered and visualized with the TIGR MeV program (http://mev.tm4.org/)37. Scatter plots of flg22-responsive genes were analyzed with Microsoft Excel. GO analysis was performed with agriGO v2.0 (www.http://systemsbiology.cau.edu.cn/agriGOv2/index.php) using default setting38.
Immunoblot analysis
Total proteins from plant materials were extracted with IP buffer (50 mM Tris at pH 7.5, 50 mM NaCl, 0.3 M sucrose, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride and 1× protease inhibitor cocktail) or 2× SDS sample buffer (100 mM Tris at pH 6.8, 4% SDS, 20% glycerol and 4% β-mercaptoethanol). After boiling with 2× SDS sample buffer, proteins were loaded on SDS-PAGE gel. Separated proteins were transferred to a nitrocellulose membrane, and probed with anti-phospho-p44/42 mitogen-activated protein kinase (Erk1/2) (Thr202/Tyr204) (Cell Signaling; 1:1,000 dilution), anti-MBP (New England Biolab; 1:2,000 dilution), anti-Myc (Cell Signaling; 1:3,000 dilution), anti-HA (Roche; 1:1,000 dilution), anti-BSL2/3 (AbFrontier; 1:2,000 dilution), anti-FLAG (Sigma; 1:1,000 dilution), anti-GST (Santa Cruz Biotechnology; 1:1,000 dilution) and anti-GFP (homemade or TRANSGENE, M018) antibodies.
Bacterial growth assays
Bacterial growth assay was performed following published procedures39. To monitor Pseudomonas syringae pathovar tomato (Pst) strain DC3000 growth in plants, leaves of 6-week-old plants were hand-infiltrated with a 1×105 colony forming unit (CFU)/mL suspension of the bacteria in 1 mM MgCl2 using a needleless syringe. For flg22 pretreatment, the leaves were hand-infiltrated with water or 1 μM flg22 one day before the bacterial inoculation. Leaf discs were collected and ground in 1 mM MgCl2 and then spotted on nutrient yeast glycerol agar plates in triplicate to determine the bacterial titer. Seedling flood inoculation assay was performed following published procedures with minor modifications40. Seedlings grown on media containing 10 μM estradiol were treated with 1 μM flg22 or distilled water one day prior to Pseudomonas inoculation. Bacteria inoculum (105 CFU/mL) containing 0.025% Silwet L-77 was dispensed into 7-day-old Arabidopsis seedlings grown on plates. Bacterial populations were evaluated from six biological replicates and each replicate represented a pooled sample of at least 20 mg of seedlings.
Yeast two-hybrid assays and in vitro pull-down assay
Yeast two-hybrid assays were performed following published procedures7. Recombinant GST-tagged proteins were purified from E. coli using Glutathione Sepharose beads (GE healthcare). The GST-tagged protein-bead complexes were incubated with 2 μg of MBP-tagged proteins in 1 mL of pull-down buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.2% NP-40 and 0.05 mg/mL BSA) at 4°C for 1 hr. Then, the protein-bead complex was washed twice with 1 mL of pull-down washing buffer (50 mM Tris, pH 7.5, 100 mM NaCl, 0.1% NP-40). Associated proteins were eluted with 2× SDS sample buffer and analyzed by immunoblotting.
In vitro kinase assays.
In vitro kinase assays using recombinant proteins were performed following published procedure6. One microgram of GST-BIK1 or GST-BIK1-Km was incubated with MBP-BSU1, MBP-BSL1, MBP-BSL2, MBP-BSL3, MBP-BSU1-Kelch, MBP-BSU1-PP or MBP-BSU1-S251A in the kinase buffer (20 mM Tris, pH 7.5, 1 mM MgCl2, 100 mM NaCl and 1 mM DTT) containing 100 μM ATP and 10 μCi [32P]-γ-ATP at 30 °C for 3 hr. For kinase assay using immunopurified BIK1, total protein was extracted from BIK1::BIK1-HA seedlings using immunoprecipitation (IP) buffer (50 mM Tris, pH 7.5, 50 mM NaCl, 0.3 M sucrose, 1% Triton X-100, 1 mM PMSF and 1× protease inhibitor cocktail), centrifuged at 1700g for 5 min. Supernatants were centrifuged at 16,000g for 10 min. The cleared supernatants were incubated with anti-HA antibody (Roche, 3F10) immobilized on Protein A/G agarose beads (Pierce) for 2 hr. The beads were then washed five times with 1 mL of IP washing buffer 1 (50 mM Tris, pH 7.5, 150 mM NaCl, 0.1% Triton X-100) followed by the kinase buffer (25 mM Tris, pH 7.5, 10 mM MgCl2 and 1 mM DTT). Aliquots of BIK1-HA-bead complex were incubated with 1 μg of MBP-BSU1 in the kinase buffer containing 10 μM ATP and 10 μCi [32P]-γ-ATP at room temperature with shaking at 1,000 rpm for 1.5 hr. Proteins were eluted with 2× SDS sample buffer and separated by SDS-PAGE. Autoradiograph images were obtained by Typhoon Trio imager (GE healthcare).
Co-immunoprecipitation assays
For transient protein co-expression in N. benthamiana, Agrobacterium (GV3101) cells containing 35S::BIK1-Myc or 35S::BSU1-YFP expression vectors were washed and resuspended in the induction medium (10 mM MES buffer at pH 5.6, 10 mM MgCl2 and 150 μM acetosyringone) and were infiltrated into young N. benthamiana leaves. Two days after infiltration, the infiltrated leaves were harvested. For the MEKK1-BSU1 interaction, Agrobacterium (GV3101) cells containing 35S::BSU1-Myc and 35S::MEKK1-YFP expression constructs were co-infiltrated in N. benthamiana leaves in the same way. Total proteins were extracted with an IP buffer. The proteins were immunoprecipitated using anti-GFP antibody-bead complexes. After washing twice with 0.5 mL of IP washing buffer 2 (50 mM Tris, pH 7.5, 50 mM NaCl, 30 mM Sucrose, 0.2% Triton X-100, 1× protease inhibitor, 0.2 mM PMSF), the co-immunoprecipitated proteins were eluted by 2× SDS sample buffer.
For experiments using Arabidopsis, 12-day old wild-type, 35S::BIK1-Myc, and 35S::BIK1-Myc;BSU1pro::BSU1-YFP seedlings were crosslinked for 15 min in 1% formaldehyde under vacuum. Total protein was extracted with an IP buffer. After centrifugation at 16,000g for 10 min, the supernatant was incubated with anti-GFP (10 μg) immobilized on UNOsphere SUPrA beads (Biorad) at 4 °C for 2.5 hr. The beads were then washed five times with 1 mL of IP washing buffer 1 and eluted samples were analyzed by immunoblot.
Quantification of flg22-induced BSU1 phosphorylation
Samples for initial screening for flg22-regulated phosphorylation sites on BSU1 were prepared as the following. Transgenic 35S::BSU1-YFP seedlings were grown on 14N (½ strength MS nutrient without nitrogen (PhytoTechnology Laboratories), NH4NO3 [0.5 g/L, Sigma], KNO3 [0.5 g/L, Sigma], pH 5.7) or 15N media (½ strength MS nutrient without nitrogen, 15NH415NO3 [0.5 g/L, Cambridge Isotope Laboratory], K15NO3 [0.5 g/L, Cambridge Isotope Laboratory], pH 5.7) for 10 days. 14N- and 15N-labeled samples were treated with 1 μM flg22 or mock solution for 10 min in reciprocal experiments. Equal amounts of tissues ground in liquid nitrogen were mixed. Proteins were then extracted in an IP buffer with a phosphatase inhibitor (Roche, PhosSTOP), centrifuged at 1700g for 5 min and filtered through miracloth, and centrifuged at 16,000g for 10 min. The supernatant was transferred to a new tube, and the same volume of IP buffer without detergent was added to dilute the detergent to 0.5%, then was incubated with anti-GFP antibody with UNOsphere SUPrA bead (Biorad) at 4 °C for 1 hr, followed by 5 times washing with wash buffer. Samples for targeted quantification of S251 phosphorylation on BSU1 were prepared as the following. The 35S::BSU1-YFP seedlings were grown on 14N or 15N media for 13 days and treated with 1 μM flg22 or mock solution for 10 min. Total proteins from mixed samples (14N-flg22/15N-mock and 14N-mock/15N-flg22) were extracted with IP buffer 2 with 1× Protease Inhibitor (Pierce) and phosphatase inhibitor (Roche, PhosSTOP), and centrifuged for 1700g for 5 min. Resulting supernatant was centrifuged at 16,000g for 10 min. The supernatant was transferred to a new tube and incubated with anti-GFP nanobody conjugated magnetic agarose beads (Allele Biotechnology) at 4 °C for 1 hr, followed by 5 times washing with IP wash buffer 4 (50 mM Tris, pH 7.5, 150 mM NaCl, 0.05% Triton X-100). Samples were eluted with 2× SDS sample buffer, separated by SDS-PAGE, followed by Colloidal Blue staining (Invitrogen).
BSU1-YFP protein bands were excised and subjected to in-gel digestion with trypsin. Peptide mixtures were desalted using C18 ZipTips (Millipore). Data were acquired in the Data Dependent Acquisition (DDA) mode. Briefly, peptides were analyzed by liquid chromatography–tandem mass spectrometry (LC-MS) on a Nanoacquity ultraperformance liquid chromatography system (Waters) connected to Linear Trap Quadrupole (LTQ) Orbitrap Velos mass spectrometer (Thermo). Peptides were separated using analytical Easy-Spray C18 columns (75 μm × 150 mm) (Thermo, ES800). The flow rate was 300 nL/min, peptides were eluted by a gradient from 2 to 30% solvent B (acetonitrile/0.1%formic acid) over 57 min, followed by a short wash at 50% solvent B. After a precursor scan was measured in the Orbitrap by scanning from mass-to-charge ratio 350 to 1400 at a resolution of 60000, the six most intense multiply charged precursors were selected for collision-induced dissociation (CID) in the linear ion trap.
MS/MS data were converted to peaklist using a script PAVA (peaklist generator that provides centroid MS2 peaklist)41, and data were searched using Protein Prospector against the TAIR database Arabidopsis thaliana from December 2010 (https://www.arabidopsis.org/), concatenated with sequence randomized versions of each protein (a total of 35386 entries). A precursor mass tolerance was set to 20 ppm and MS/MS2 tolerance was set to 0.6 Da. Carbamidomethylcysteine was searched as a constant modification. Variable modifications include protein N-terminal acetylation, peptide N-terminal Gln conversion to pyroglutamate, and Met oxidation. Subsequent searches were performed to find those peptides modified by phosphorylation. The search parameters were as above, but this time allowing for phosphorylation on serine, threonine, and tyrosine. Assignment of modified peptide was checked manually and is illustrated in Fig. 3. 15N-labeled searches were done the same as mentioned above, considering all 20 amino acids are constantly modified by 15N labeling. FDR 1% was set for both proteins and peptides. For Quantification, 15N labeling efficiency was manually checked. “15N labeling” was chosen as a quantitative method using Protein Prospector with automatic adjustment of L:H intensity ratios with labeling efficiency.
For targeted quantification, data was acquired in the Parallel Reaction Monitoring (PRM) mode. Peptides were separated using an EasyLC1200 system (Thermo) connected to high performance quadrupole Orbitrap mass spectrometer Q Exactive HF (Thermo). Peptides were separated using analytical Easy-Spray C18 columns (75 μm × 250 mm) (Thermo, ES802). The flow rate was 300 nL/min, peptides were eluted by a gradient from 3 to 28% solvent B (80% acetonitrile/0.1% formic acid) over 45 min, 28 to 44% solvent B for 15 min, followed by a short wash at 90% solvent B. Samples were analyzed using PRM mode with an isolation window of 1.4 Th. PRM scans were done using 30000 resolutions (AGC target 2e5, 200 ms maximum injection time) triggered by an inclusion list (Supplementary Table 5). Normalized collision energy 27 was used in a higher-energy dissociation mode (HCD). PRM data were analyzed using skyline software42.
Quantitative real-time PCR
Total RNA was extracted from seedlings using the Spectrum Plant Total RNA kit (Sigma). M-MLV reverse transcriptase (Fermentas) was used to synthesize complementary DNA (cDNA) from the RNA. Quantitative real-time PCR (qPCR) was carried out using LightCycler 480 (Roche) and the SYBR Green Master Mix (Bioline). FRK1 and At2g17740 primers were previously described43. ROF2 and CRK13 primers are listed in Supplementary Table 6. PP2A was used as an internal reference44.
Extended Data
Extended Data Fig. 1 |. Gene ontology (GO) analysis of genes differentially expressed in bsu-q.
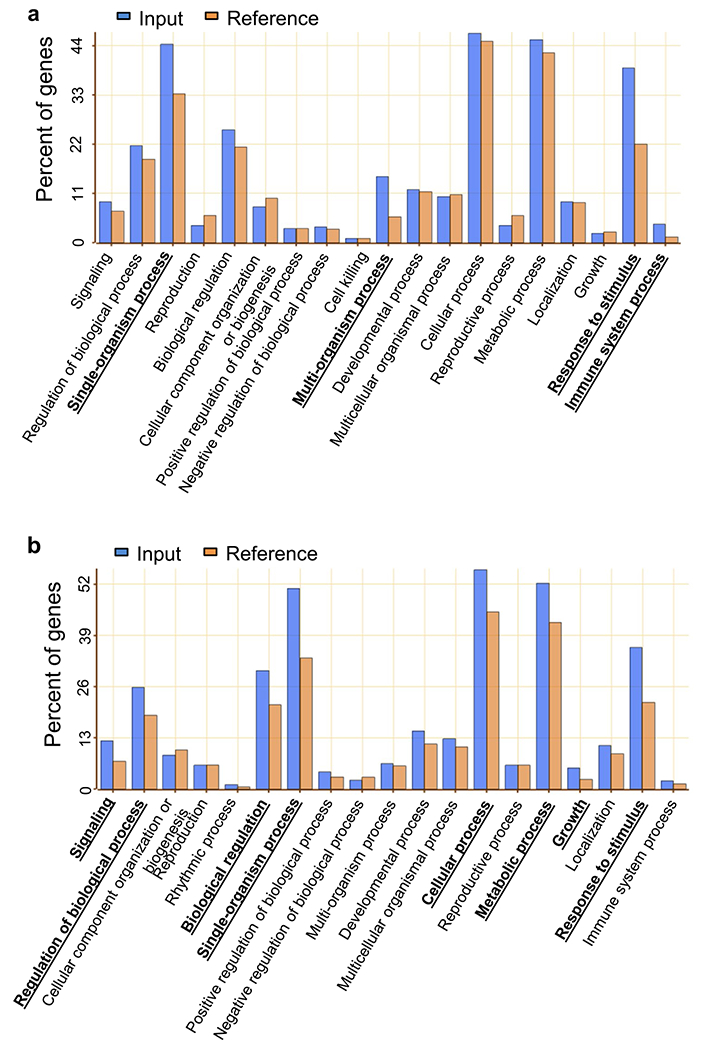
(a, b) Enrichment of GO annotation in categories of biological process for genes increased (a) and decreased (b) in bsu-q. Blue and orange columns indicate the percent of genes of input (Supplementary Table 2) and reference (TAIR10), respectively. Bold with underlines indicate enriched GO categories of biological process.
Extended Data Fig. 2 |. Expression of flagellin-induced genes are altered in bsu-q.
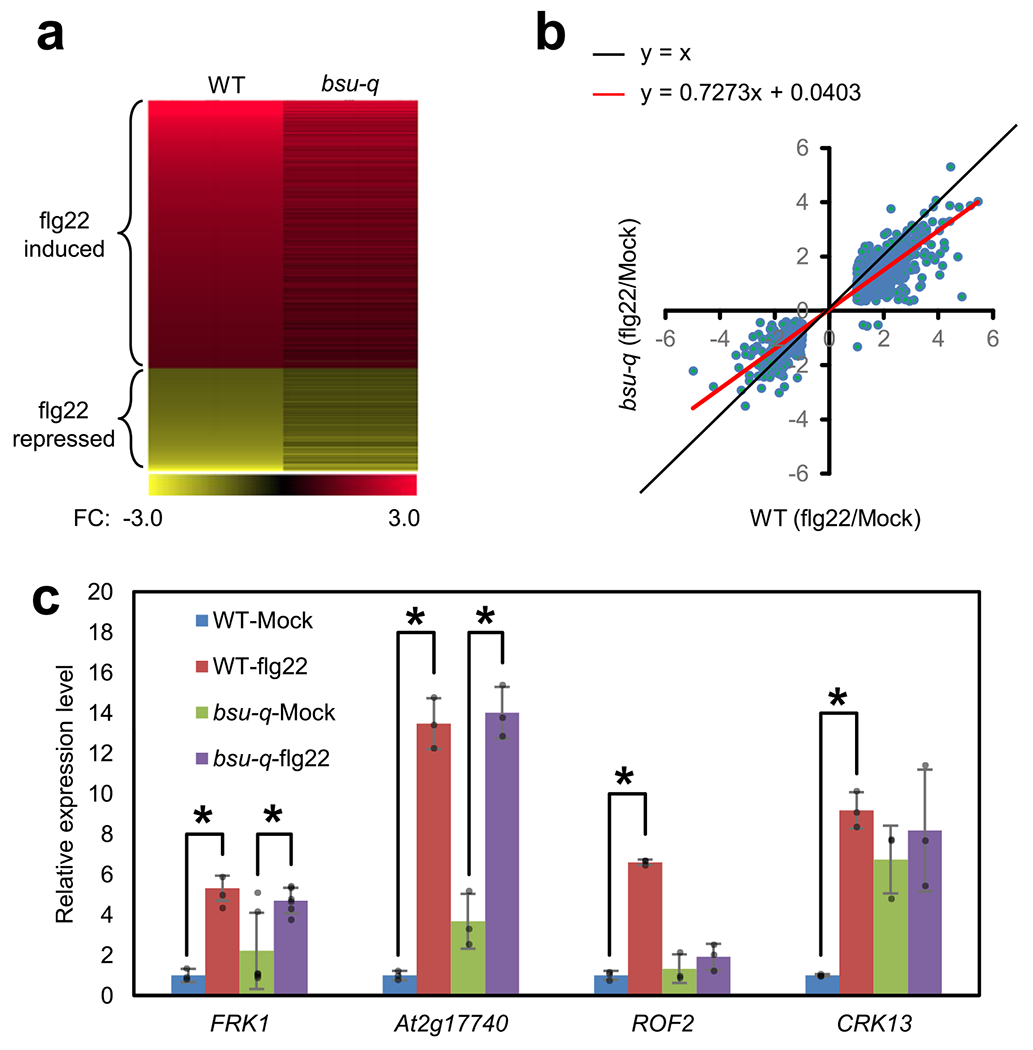
(a, b) The bsu-q mutant has defects in flg22-induced gene expression. Heat map (a) and scatter plot (b) of log2 fold change (FC, flg22/mock) values of flg22-responsive genes in wild type (WT) and bsu-q from RNA-seq data. (c) Quantitative RT-PCR of flg22-induced FRK1, At2g17740, ROF2 and CRK13 expression in WT and bsu-q. The relative expression levels are normalized to the PP2A gene expression and to the wild type sample. Asterisks indicate statistically significant difference between mock and flg22 treated samples (two-tail t-test, p = 6.3014E-7, 0.02168, 0.002650, 0.0006797, 1.9289E-5 and 0.003883 from left to right). The graph shows average expression level ± SEM of six biological repeats for FRK1 (n = 6) and three biological repeats for other genes (n = 3).
Extended Data Fig. 3 |. Loss of BSU1 family impairs flg22 induction of ROS and increases SA level.
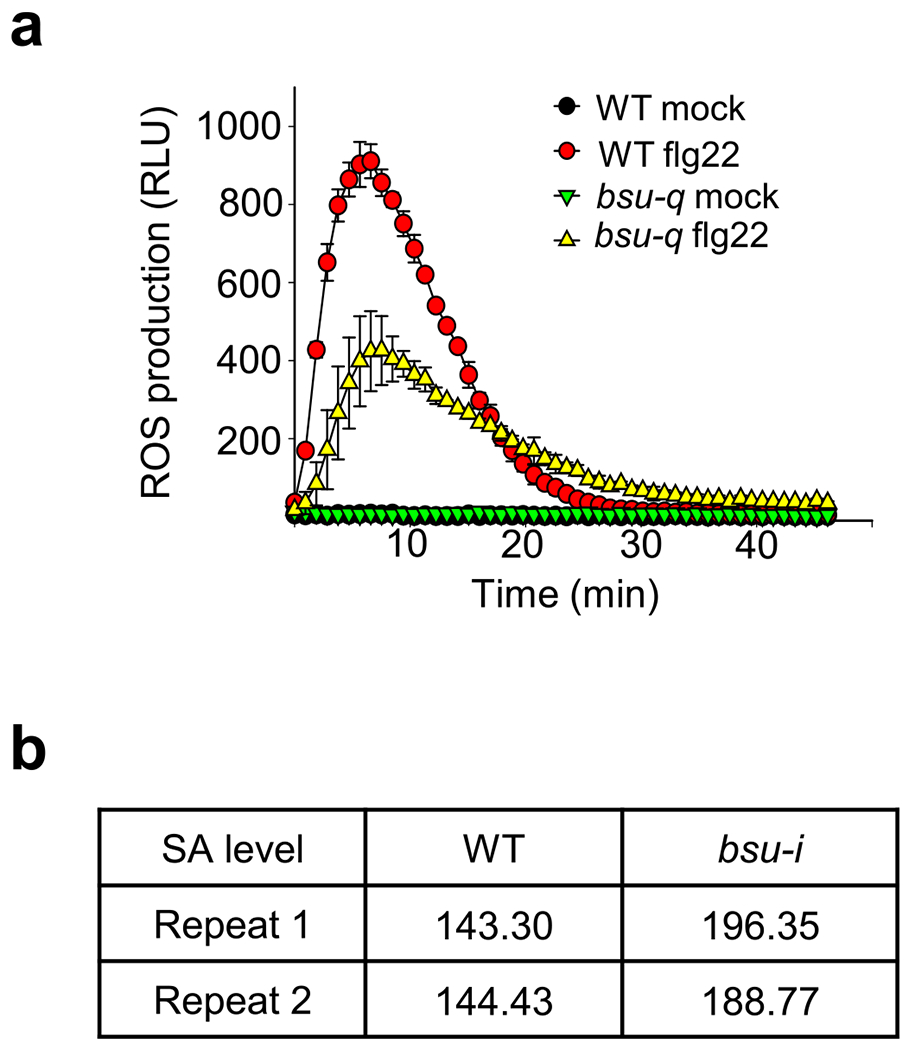
(a) Flg22-induced ROS burst in WT and bsu-q. are indicated. Each data point represents the average of relative light unit (RLU) ± SEM of at least 12 replicates from 2 biological repeats (6 samples for each repeat). Each repeat showed similar results. (b) The bsu-i mutant accumulates increased levels of salicylic acid. The SA levels (microgram per gram fresh tissue) of wild type and bsu-i were measured from 0.1 g of 11-day-old seedlings grown on 10 μM estradiol in two biological repeats.
Extended Data Fig. 4 |. The flg22- and elf18-induced MPK3/6 phosphorylation in the BSU1 family mutants.
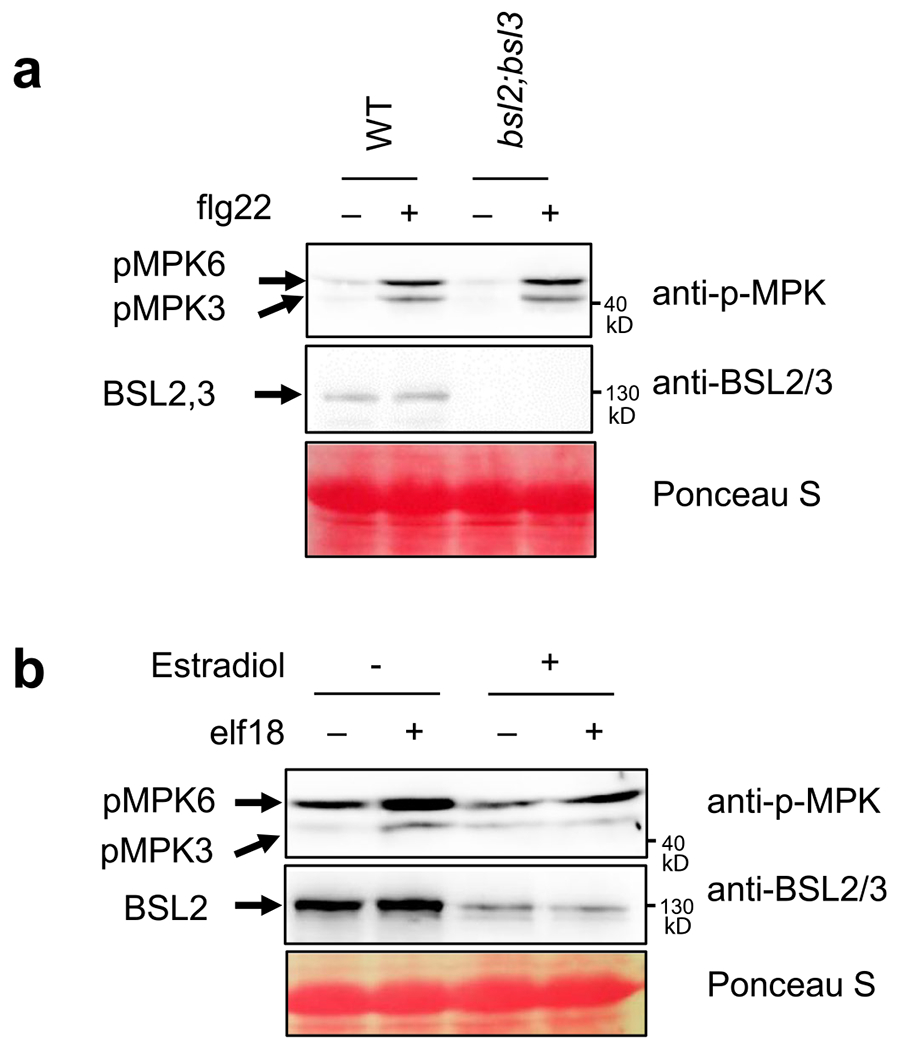
(a) Flg22-induced MPK3/6 phosphorylation in wild-type (WT) and bsl2;bsl3. Ten-day-old seedlings were treated with 100 nM flg22 for 10 min. Immunoblot was performed using anti-phospho-p44/42 MAPK and anti-BSL2/3 antibodies. Ponceau S staining shows equal protein loading. (b) Elf18-induced MPK3/6 phosphorylation in bsu-i. Twelve-day-old seedlings grown on 20 μM estradiol containing media were treated with 1 μM elf18 for 10 min. Immunoblot was performed using anti-phospho-p44/42 MAPK and anti-BSL2/3 antibodies. Ponceau S staining shows equal protein loading.
Extended Data Fig. 5 |. Yeast-two-hybrid assays for interactions between BIK1 family RLCKs and BSU1/BSLs proteins.
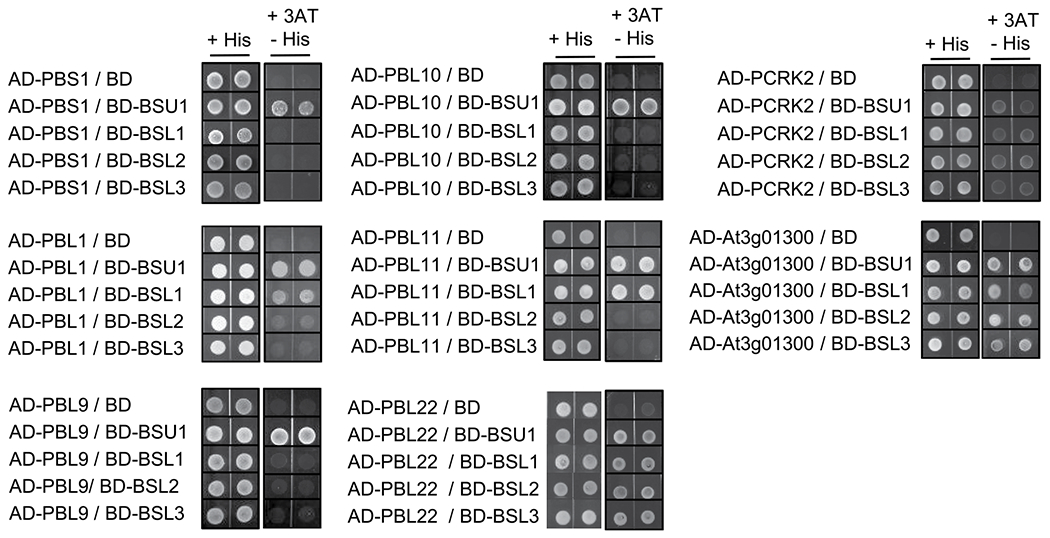
Yeast-two-hybrid assay for interaction between BIK1/PBLs and BSU1/BSLs proteins. Yeast cells co-expressing AD-BIK1/PBLs and BD-BSU1/BSLs were grown on synthetic dropout medium containing 2.5 mM 3-amino-1, 2, 4-triazole (3AT).
Extended Data Fig. 6 |. SILIA IP-MS PRM analysis of flg22-induced BSU1 phosphorylation.
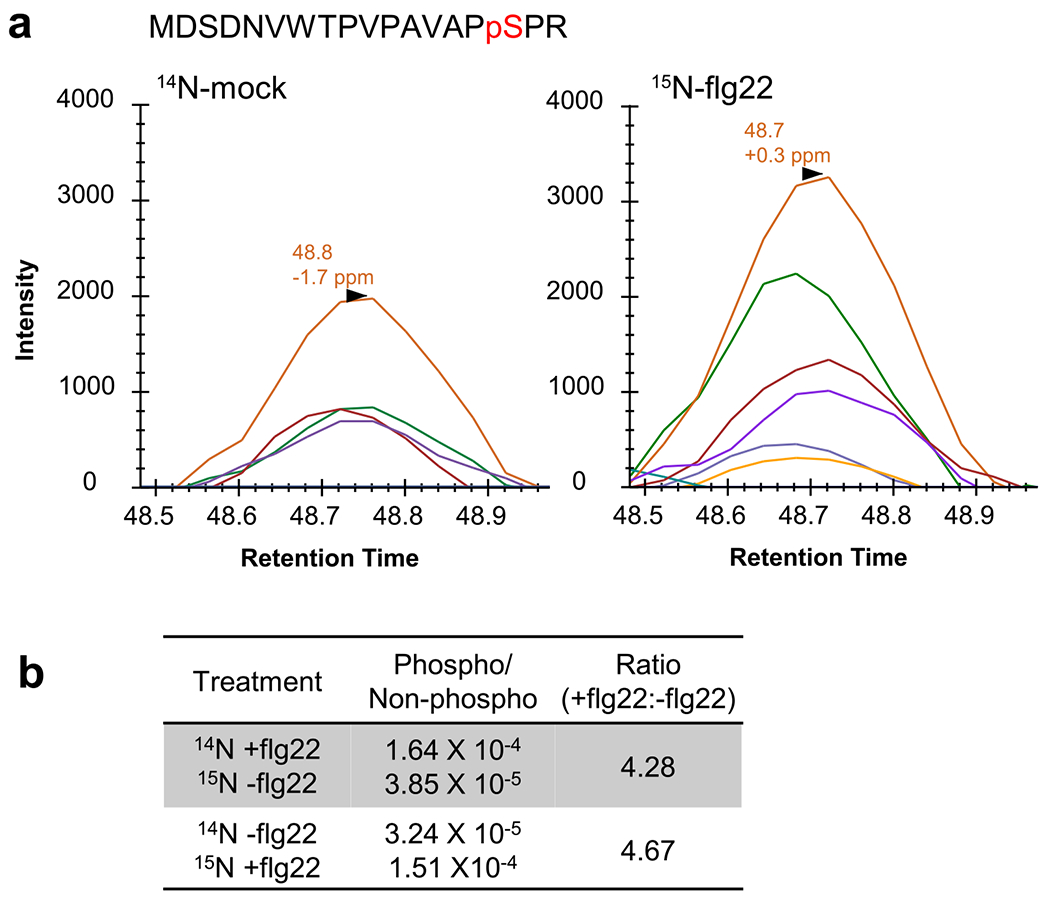
(a) Chromatograms of fragment ions from the replicate SILIA IP-MS PRM experiment of Fig. 3c using reciprocal labeling. (b) The ratio of phosphorylated and non-phosphorylated MDSDNVWTPVPAVAPpSPR peptides. The values of peak area were used for quantification. The ratios of S251 phosphorylation between flg22-treated (+flg22) and mock-treated (−flg22) samples are shown.
Extended Data Fig. 7 |. Alignment of sequence around S251 of BSU1 with those of paralogs.
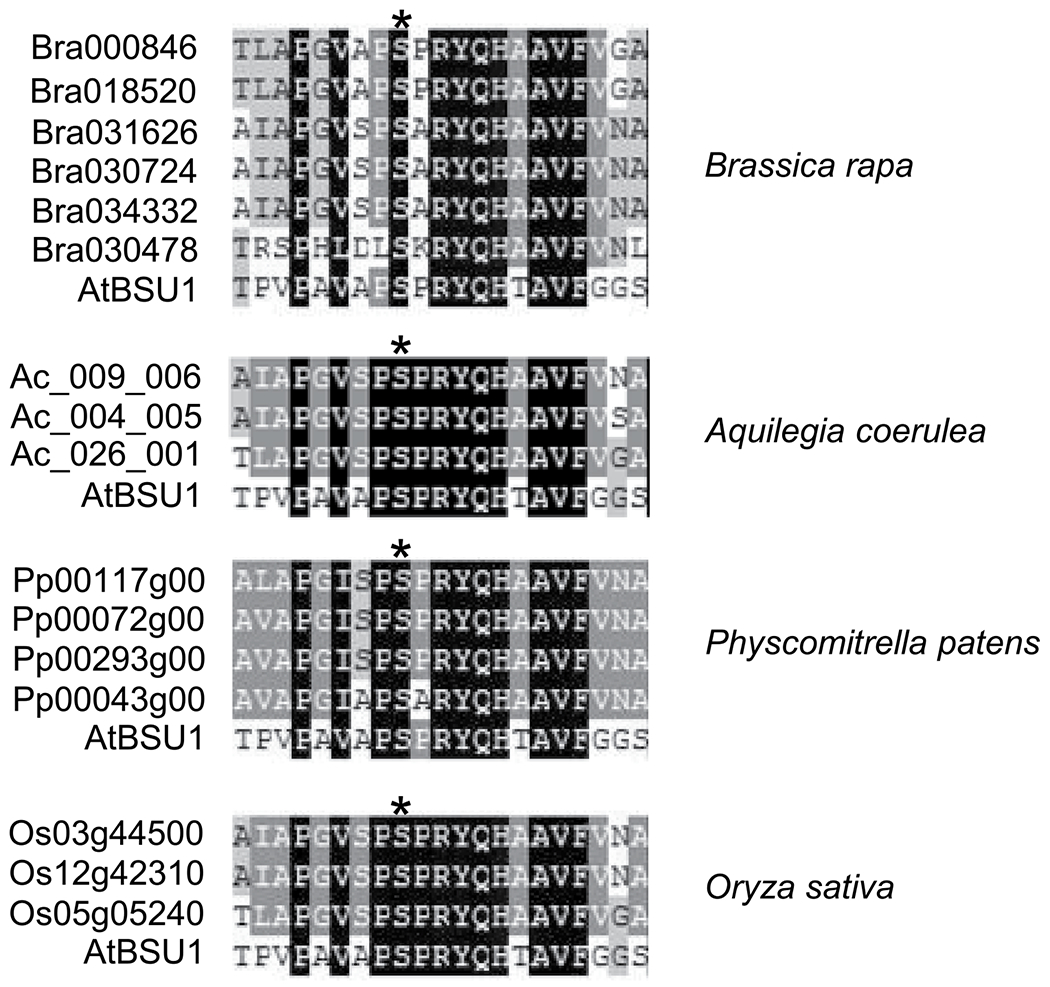
Amino acid sequences of the indicated species were obtained from Maselli et al., 2014 (22). Asterisks indicate BSU1 S251.
Extended Data Fig. 8 |. Relative band intensity of Fig. 5a, c.
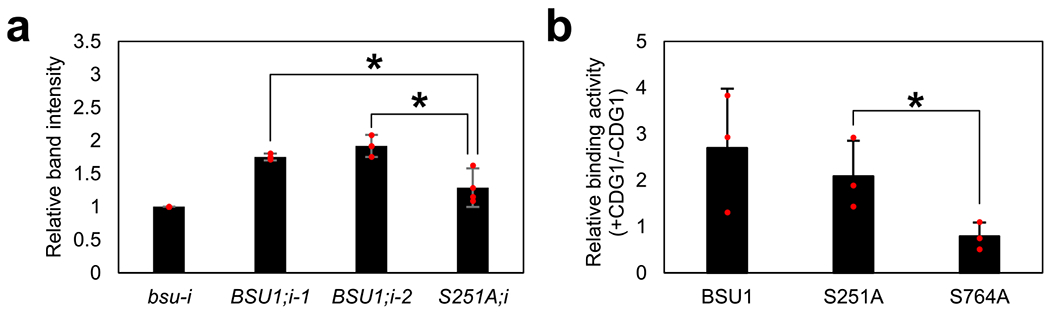
(a) Relative band intensities of immunoblot images of flg22-treated samples using anti-phospho-p44/42 MAPK antibody (anti-p-MPK) in Fig. 5a were measured by ImageJ. The band intensity of each bsu-i sample was set to 1. The graph shows the average band intensity ± SEM from three repeat experiments. One-tail t-tests show that the band intensities of BSU1;i-1 and BSU1;i-2 are significantly stronger than that of S251A;i (p = 0.0268 and p = 0.0157). (b) Band intensities of immunoblot images, represented by Fig. 5c, were measured by ImageJ. The band intensity of GST-BIN2 was divided by that of MBP-BSU1, and then the ratio between +GST-CDG1 and −GST-CDG1 is presented to show the effect of CDG1 on BSU1-BIN2 binding. The graph shows the average binding activity ± SEM from three repeat experiments. One-tail t-test shows that the binding activity of S251A is significantly higher than that of S764A. (p = 0.041983).
Extended Data Fig. 9 |. BSU1 protein level of seedlings in Fig. 5d, e.
Total protein was extracted from 7-day old light-grown seedlings. BSU1-YFP protein level was analyzed by immunoblotting using anti-GFP antibody. Ponceau S staining image shows equal protein loading. These transgenic lines were used in Fig. 5d, e.
Supplementary Material
Supplementary Table 1. Genes differentially expressed in bsu-q vs. wild type (Col-0).
RNA-seq data of genes that are differentially expressed in bsu-q compared to wild type with fold change >2 (UP: bsu-q/Col-0; DOWN: Col-0/bsu-q) and q-value<0.05.
Supplementary Table 2. GO enrichment analysis of genes differentially expressed in bsu-q.
Supplementary Table 3. RNA-seq analysis of flg22 responsive genes in wild type and bsu-q. RNA-seq analysis of wild-type (a) and bsu-q (b) plants 1 hr after treatment with 1 μM flg22 or mock.
Supplemental Table 4: RNA-seq data. Fragments per kilobase per million read (FPKM) is shown for wildtype (WT) and bsu-q (bsuq) treated with mock (ct) or flg22 (fg).
Supplementary Table 5. Inclusion list for S251 PRM.
Supplementary Table 6. Oligos used in this study.
Acknowledgements
We thank Jian-Min Zhou for sharing BIK1::BIK1-HA and MEKK1-FLAG seeds, Libo Shan and Ping He for GST-BIK1 and GST-BIK1Km constructs. We thank Andres Valentino Reyes for generating the bee swam box plot figures. This research was supported by grants from NIH (R01GM066258 to Z-Y.W.), National Research Foundation of Korea funded by the Ministry of Science, ICT, Future Planning (NRF-2021R1A2C1006617 and 2020R1A6A1A06046728 to T.W.K., 2021R1A2C1007516 to S-K.K.), NSF (NSF-IOS 2026368 to M.B.M.), NIH (R01GM135706 to S.L.X.), by Howard Hughes Medical Institute and NIH (P41GM103481 to A.L.B.), and Carnegie Institution for Science endowment fund to the Carnegie mass spectrometry facility.
Footnotes
Competing interests:
The authors declare no competing interests.
Supplementary Information is available for this paper.
Data availability:
The RNA-seq data that support the findings of this study have been deposited in GEO with the accession code GSE140037. The mass spectrometry proteomics data are available via ProteomeXchange with identifier PXD016283 (Username: reviewer67695@ebi.ac.uk; Password: pCDR9H2R, for DDA mode) and PXD016257 (Username: reviewer88035@ebi.ac.uk; Password: IQKH8jha, for PRM mode).
REFERENCES
- 1.Shiu SH et al. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16, 1220–1234, doi: 10.1105/tpc.020834 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li J et al. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110, 213–222, doi: 10.1016/s0092-8674(02)00812-7 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Nam KH & Li J BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110, 203–212 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Chinchilla D et al. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448, 497–500, doi: 10.1038/nature05999 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Tang W et al. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321, 557–560, doi: 10.1126/science.1156973 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim TW, Guan SH, Burlingame AL & Wang ZY The CDG1 Kinase Mediates Brassinosteroid Signal Transduction from BRI1 Receptor Kinase to BSU1 Phosphatase and GSK3-like Kinase BIN2. Mol Cell 43, 561–571, doi: 10.1016/j.molcel.2011.05.037 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim TW et al. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat Cell Biol 11, 1254–U1233, doi: 10.1038/ncb1970 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang WQ et al. PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat Cell Biol 13, 124–U149, doi: 10.1038/ncb2151 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim TW & Wang ZY Brassinosteroid Signal Transduction from Receptor Kinases to Transcription Factors. Annu Rev Plant Biol 61, 681–704, doi: 10.1146/annurev.arplant.043008.092057 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Lu D et al. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci U S A 107, 496–501, doi: 10.1073/pnas.0909705107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J et al. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7, 290–301, doi: 10.1016/j.chom.2010.03.007 (2010). [DOI] [PubMed] [Google Scholar]
- 12.DeFalco TA & Zipfel C Molecular mechanisms of early plant pattern-triggered immune signaling. Mol Cell 81, 3449–3467, doi: 10.1016/j.molcel.2021.07.029 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Lin W et al. Inverse modulation of plant immune and brassinosteroid signaling pathways by the receptor-like cytoplasmic kinase BIK1. Proc Natl Acad Sci U S A 110, 12114–12119, doi: 10.1073/pnas.1302154110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi H et al. BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis. Plant Cell 25, 1143–1157, doi: 10.1105/tpc.112.107904 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naveed ZA, Wei X, Chen J, Mubeen H & Ali GS The PTI to ETI Continuum in Phytophthora-Plant Interactions. Front Plant Sci 11, 593905, doi: 10.3389/fpls.2020.593905 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ngou BPM, Ahn HK, Ding P & Jones JDG Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 592, 110–115, doi: 10.1038/s41586-021-03315-7 (2021). [DOI] [PubMed] [Google Scholar]
- 17.Yuan M et al. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 592, 105–109, doi: 10.1038/s41586-021-03316-6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhai K et al. NLRs guard metabolism to coordinate pattern- and effector-triggered immunity. Nature, doi: 10.1038/s41586-021-04219-2 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Wu YJ et al. Loss of the common immune coreceptor BAK1 leads to NLR-dependent cell death. P Natl Acad Sci USA 117, 27044–27053, doi: 10.1073/pnas.1915339117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Veronese P et al. The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 18, 257–273, doi: 10.1105/tpc.105.035576 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mora-Garcia S et al. Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev 18, 448–460, doi: 10.1101/gad.1174204 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maselli GA et al. Revisiting the evolutionary history and roles of protein phosphatases with Kelch-like domains in plants. Plant Physiol 164, 1527–1541, doi: 10.1104/pp.113.233627 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim TW, Michniewicz M, Bergmann DC & Wang ZY Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482, 419–422, doi: 10.1038/nature10794 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turnbull D et al. AVR2 Targets BSL Family Members, Which Act as Susceptibility Factors to Suppress Host Immunity. Plant Physiol 180, 571–581, doi: 10.1104/pp.18.01143 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zipfel C et al. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125, 749–760, doi: 10.1016/j.cell.2006.03.037 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Asai T et al. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983, doi: 10.1038/415977a (2002). [DOI] [PubMed] [Google Scholar]
- 27.Bi G et al. Receptor-Like Cytoplasmic Kinases Directly Link Diverse Pattern Recognition Receptors to the Activation of Mitogen-Activated Protein Kinase Cascades in Arabidopsis. Plant Cell 30, 1543–1561, doi: 10.1105/tpc.17.00981 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kadota Y et al. Direct Regulation of the NADPH Oxidase RBOHD by the PRR-Associated Kinase BIK1 during Plant Immunity. Mol Cell 54, 43–55, doi: 10.1016/j.molcel.2014.02.021 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Kong Q et al. The MEKK1-MKK1/MKK2-MPK4 kinase cascade negatively regulates immunity mediated by a mitogen-activated protein kinase kinase kinase in Arabidopsis. Plant Cell 24, 2225–2236, doi: 10.1105/tpc.112.097253 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lukowitz W, Roeder A, Parmenter D & Somerville C A MAPKK kinase gene regulates extra-embryonic cell fate in Arabidopsis. Cell 116, 109–119, doi: 10.1016/s0092-8674(03)01067-5 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Guo X, Park CH, Wang ZY, Nickels BE & Dong J A spatiotemporal molecular switch governs plant asymmetric cell division. Nat Plants 7, 667–680, doi: 10.1038/s41477-021-00906-0 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim EJ et al. Oligomerization between BSU1 Family Members Potentiates Brassinosteroid Signaling in Arabidopsis. Mol Plant 9, 178–181, doi: 10.1016/j.molp.2015.09.012 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Curtis MD & Grossniklaus U A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133, 462–469, doi: 10.1104/pp.103.027979 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hartwig T et al. Propiconazole Is a Specific and Accessible Brassinosteroid (BR) Biosynthesis Inhibitor for Arabidopsis and Maize. Plos One 7, doi:ARTN e36625 10.1371/journal.pone.0036625 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Earley KW et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant Journal 45, 616–629, doi: 10.1111/j.1365-313X.2005.02617.x (2006). [DOI] [PubMed] [Google Scholar]
- 36.Goff SA et al. The iPlant collaborative: cyberinfrastructure for plant biology. Front Plant Sci 2, doi:ARTN 3410.3389/fpls.2011.00034 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saeed AI et al. TM4: A free, open-source system for microarray data management and analysis. Biotechniques 34, 374-+, doi:DOI 10.2144/03342mt01 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Tian T et al. agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res 45, W122–W129, doi: 10.1093/nar/gkx382 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fan M et al. The bHLH Transcription Factor HBI1 Mediates the Trade-Off between Growth and Pathogen-Associated Molecular Pattern-Triggered Immunity in Arabidopsis. Plant Cell 26, 828–841, doi: 10.1105/tpc.113.121111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishiga Y, Ishiga T, Uppalapati SR & Mysore KS Arabidopsis seedling flood-inoculation technique: a rapid and reliable assay for studying plant-bacterial interactions. Plant Methods 7, doi:Artn 32 10.1186/1746-4811-7-32 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guan SH, Price JC, Prusiner SB, Ghaemmaghami S & Burlingame AL A Data Processing Pipeline for Mammalian Proteome Dynamics Studies Using Stable Isotope Metabolic Labeling. Mol Cell Proteomics 10, doi:ARTN M111.01072810.1074/mcp.M111.010728 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.MacLean B et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968, doi: 10.1093/bioinformatics/btq054 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He P et al. Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell 125, 563–575, doi: 10.1016/j.cell.2006.02.047 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Oh E, Zhu JY & Wang ZY Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol 14, 802–U864, doi: 10.1038/ncb2545 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Genes differentially expressed in bsu-q vs. wild type (Col-0).
RNA-seq data of genes that are differentially expressed in bsu-q compared to wild type with fold change >2 (UP: bsu-q/Col-0; DOWN: Col-0/bsu-q) and q-value<0.05.
Supplementary Table 2. GO enrichment analysis of genes differentially expressed in bsu-q.
Supplementary Table 3. RNA-seq analysis of flg22 responsive genes in wild type and bsu-q. RNA-seq analysis of wild-type (a) and bsu-q (b) plants 1 hr after treatment with 1 μM flg22 or mock.
Supplemental Table 4: RNA-seq data. Fragments per kilobase per million read (FPKM) is shown for wildtype (WT) and bsu-q (bsuq) treated with mock (ct) or flg22 (fg).
Supplementary Table 5. Inclusion list for S251 PRM.
Supplementary Table 6. Oligos used in this study.
Data Availability Statement
The RNA-seq data that support the findings of this study have been deposited in GEO with the accession code GSE140037. The mass spectrometry proteomics data are available via ProteomeXchange with identifier PXD016283 (Username: reviewer67695@ebi.ac.uk; Password: pCDR9H2R, for DDA mode) and PXD016257 (Username: reviewer88035@ebi.ac.uk; Password: IQKH8jha, for PRM mode).


