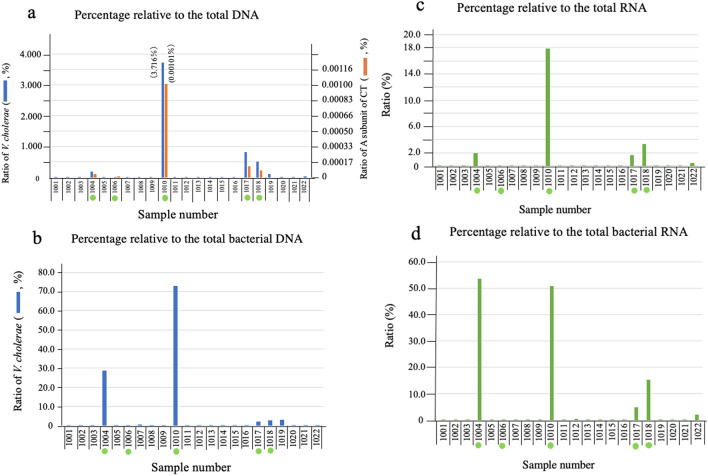
Figure 6.
The ratio of DNA and RNA derived from V. cholerae in stool samples of the specific-pathogen-free patients. The stool samples from 22 diarrheal patients in which no etiological agent of diarrhea, including V. cholerae, was detected by our bacterial examination in the laboratory were analyzed in this examination. The extraction of DNA and RNA, and the preparation of reverse-transcribed DNA samples from the RNA samples were performed in the same manner as in Fig. 2. The origin of the reads obtained in this analysis was assigned by mapping to a database that included human and microorganism sequences. The obtained numbers of total reads, total bacterial reads, reads originating from V. cholerae, reads from ctxA in each sample are shown in Supplementary Tables S5 (the data from DNA) and S6 (the data from RNA). The percentages of reads of DNA from V. cholerae (blue bar in a) and of reads of DNA from ctxA (red bar in a) relative to the total DNA reads, and the percentages of reads of DNA from V. cholerae relative to the total bacterial DNA reads (b) are presented. Similarly, the results obtained from the RNA samples are presented in (c) and (d). The (c) and (d) show the percentages of reads of RNA from V. cholerae relative to the total RNA reads and to the reads of total bacterial RNA, respectively. The samples indicated by green circles are the samples of interest in this manuscript, as described in the text.