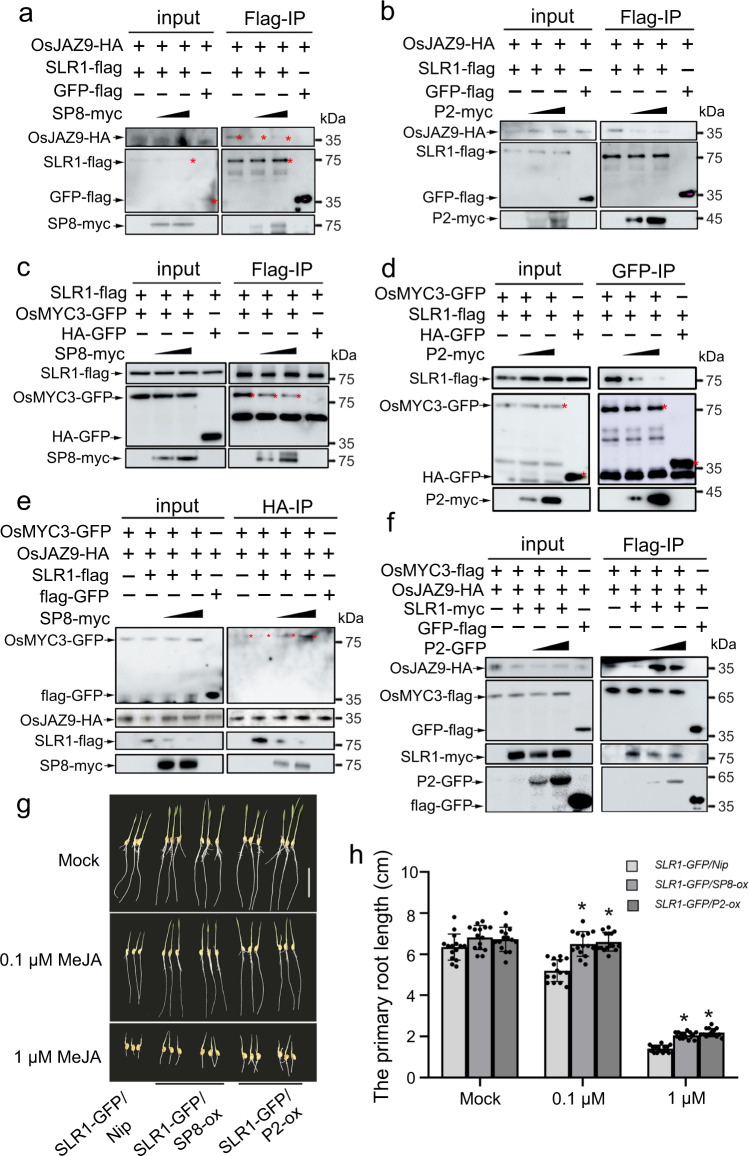
Fig. 6. Viral proteins restrict SLR1 to activate JA antiviral signaling.
a, b Protein competition analyzed by Co-IP assays in vivo. OsJAZ9-HA and SLR1-flag were infiltrated using Agrobacterium together with increasing amounts of SP8-myc (a) or P2-myc (b) in leaves of N. benthamiana, GFP-flag serves as negative control. Cultures were pelleted to a final OD600 of 0.5, increasing amounts of SP8 and P2 following agrobacterium infection with final OD600 = 0.5 or OD600 = 1.0, respectively. The co-infiltrated leaves were treated with MG132 (50 µM) or DMSO at 24 hpi and then harvested 24 h later for coimmunoprecipitation with Flag-tag paramagnetic beads. Each experiment was repeated three times with similar results. c, d Protein competition analyzed by Co-IP assays in vivo. OsMYC3 and SLR1 were infiltrated using Agrobacterium together with increasing amount of SP8-myc (c) or P2-myc (d) in leaves of N. benthamiana, HA-GFP used as negative controls. Cultures were pelleted to a final OD600 of 0.5, increasing amounts of SP8 and P2 following agrobacterium infection with final OD600 = 0.5 or OD600 = 1.0, respectively. The co-infiltrated leaves were treated with MG132 (50 µM) or DMSO at 24 hpi and then harvested 24 h later for coimmunoprecipitation with Flag-tag paramagnetic beads. Each experiment was repeated three times with similar results. e, f. Protein competition analyzed by Co-IP assays in vivo. OsMYC3-GFP and OsJAZ9-HA were infiltrated using Agrobacterium together with SLR1-flag in leaves of N. benthamiana. Increasing amounts of SP8-myc (e) or (f) were mixed to degrade SLR1 and GFP-flag was used as negative control. Cultures were pelleted to a final OD600 of 0.5, increasing amounts of SP8 and P2 following agrobacterium infection with final OD600 = 0.5 or OD600 = 1.0, respectively. Proteins were harvested at 48 hpi and then immunoprecipitated with corresponding HA-tag (e) or Flag-tag (f) paramagnetic beads. Each experiment was repeated three times with similar results. g Phenotypes of SLR1-GFP/Nip, SLR1-GFP/SP8-ox and SLR1-GFP/P2-ox seedlings grown for 7 days on rice nutrient solutions with different concentrations of MeJA (0, 0.1, 1 µM), n = 3 biologically independent replicates per genotype. All images were photographed using a digital camera. Scale bar = 3 cm. h Root lengths of SLR1-GFP/Nip, SLR1-GFP/SP8-ox and SLR1-GFP/P2-ox seedlings. Error bars represent SD, * at the top of columns indicate significant differences (p < 0.05) based on Fisher’s least significant difference tests. Values were obtained from n = 15 biologically independent plants. Source data including uncropped scans of gels (a–f) and p values of statistic tests (h) are provided in the Source data file.