Summary
mTORC1 is aberrantly activated in cancer and in the genetic tumor syndrome tuberous sclerosis complex (TSC), which is caused by loss-of-function mutations in the TSC complex, a negative regulator of mTORC1. Clinically approved mTORC1 inhibitors, such as rapamycin, elicit a cytostatic effect that fails to eliminate tumors and is rapidly reversible. We sought to determine the effects of mTORC1 on the core regulators of intrinsic apoptosis. In TSC2-deficient cells and tumors, we find that mTORC1 inhibitors shift cellular dependence from MCL-1 to BCL-2 and BCL-XL for survival, thereby altering susceptibility to BH3 mimetics that target specific pro-survival BCL-2 proteins. The BCL-2/BCL-XL inhibitor ABT-263 synergizes with rapamycin to induce apoptosis in TSC-deficient cells and in a mouse tumor model of TSC, resulting in a more complete and durable response. These data expose a therapeutic vulnerability in regulation of the apoptotic machinery downstream of mTORC1 that promotes a cytotoxic response to rapamycin.
Subject areas: Biological sciences, Cancer, Cell biology, Molecular biology
Graphical abstract

Highlights
-
•
mTORC1 inhibition alters expression of BCL-2 family proteins in TSC-deficient cells
-
•
mTORC1 inhibitors shift cellular dependence from MCL-1 to BCL-2/BCL-XL for survival
-
•
ABT-263 synergizes with mTORC1 inhibitors in TSC-deficient mouse and human cells
-
•
ABT-263 improves the anti-tumor durability of rapamycin in a TSC tumor model
Biological sciences; Cancer; Cell biology; Molecular biology
Introduction
The mechanistic target of rapamycin (mTOR) complex 1 (mTORC1) is a critical convergence point for the most common oncogenic signals, leading to aberrant, growth factor-independent activation of mTORC1 signaling in the majority of human cancers.1,2,3 The activation of mTORC1 drives growth-promoting anabolic processes, including a coordinated increase in the synthesis of proteins, lipids and nucleotides.4 The tuberous sclerosis complex (TSC) protein complex, comprising TSC1, TSC2 and TBC1D7, is an essential negative regulator of mTORC1. Although TSC1 and TSC2 themselves are tumor suppressors, the TSC complex is often functionally inactivated in cancer through mutations in upstream oncogenes and tumor suppressors, thus providing the most frequent route of mTORC1 activation in cancer. The genetic tumor syndrome TSC results from germline or mosaic loss-of-function mutations in TSC1 or TSC2, leading to constitutive activation of mTORC1 in the widespread neoplastic lesions inflicting individuals with TSC. This autosomal dominant disorder is characterized by tumors in multiple organ systems, most frequently affecting the brain, kidney, skin, heart and lung, as well as neurological manifestations, including epilepsy and autism spectrum disorder.5,6TSC1 and TSC2 mutations can also occur in sporadic cancers, with the most common being bladder and liver cancer.7,8,9
Owing to the prevalence of mTORC1 activation in tumors, there has been great interest in targeting mTORC1 pharmacologically. However, mTORC1 inhibitors have had limited success as single agents.10 The allosteric mTORC1 inhibitor rapamycin (also known as sirolimus or Rapamune) and its analogs (e.g., everolimus, temsirolimus), collectively referred to as rapalogs, are partial inhibitors of mTORC1 and currently the only clinically approved mTORC1 inhibitors for any indication.11,12 Inhibitors targeting the mTOR kinase domain, such as MLN0128, CC-223, and AZD-2014, have been used in many preclinical studies and are currently being tested to treat cancers in Phase I/II clinical trials (clinicaltrials.gov). Although these compounds lead to stronger inhibition of mTORC1, they also fully suppress mTOR complex 2 (mTORC2), a second physically and functionally distinct mTOR-containing protein complex. mTOR kinase domain inhibitors hold promise as future cancer therapeutics but are generally more toxic and pharmacologically inferior to rapalogs.13,14,15 Clinical evidence indicates that rapalogs are effective in treating TSC-associated lesions, including renal angiomyolipoma (AML), subependymal giant cell astrocytoma (SEGA), and pulmonary lymphangioleiomyomatosis (LAM).16,17,18 However, even in these settings where tumor growth is driven specifically by mTORC1 activation, rapalogs only partially shrink tumors, without eradicating them.11,19 Importantly, in both TSC mouse models and TSC and LAM patients, tumors exhibit rapid regrowth on halting rapalog treatment, suggesting a lack of cytotoxicity within the tumors and a prompt restoration of mTORC1-driven growth.5,20,21,22 In cancers with loss of function mutations in TSC1 or TSC2, rapalogs also cause growth-inhibitory effects, but likewise remain cytostatic compounds.7,8
Although much progress has been made to advance our understanding of mTOR signaling, the effects of mTORC1 activation and inhibition on cell-intrinsic survival and death mechanisms are not fully understood. BCL-2 family proteins are the key cellular regulators of apoptosis, the most common form of cell death. The coordinated actions of these proteins result in either prevention or stimulation of mitochondrial outer membrane permeabilization (MOMP) to respectively promote cell survival or induce apoptotic cell death.23 MOMP releases cytochrome c resulting in the irreversible activation of cytosolic caspases that cause cell death. Pro-apoptotic proteins can be subdivided into effector proteins (BAK and BAX) which form the MOMP pore, activator proteins (BIM and BID) which bind to and activate the effector proteins, and sensitizer proteins (PUMA, NOXA, BIK, BAD, BMF) which bind to and inhibit pro-survival proteins. The group of pro-survival proteins (MCL-1, BCL-2, BCL- XL, BCL-W, and BFL-1/A1) act by engaging and inhibiting the activator or effector proteins.24 Changes in the cellular abundance and stoichiometry of BCL-2 family proteins has been implicated in the pathogenesis of many cancers, most notably in hematological malignancies, leading to the development of therapies that target the pro-survival BCL-2 family proteins.25
BH3 mimetics are synthetic small molecules that mimic specific pro-apoptotic proteins and can sequester specific pro-survival BCL-2 proteins, promoting apoptosis in cells dependent on those pro-survival proteins.26 Although BH3 mimetics have shown efficacy as single agents, this is generally limited to hematological malignancies.27,28,29 However, BH3 mimetics may also be used to enhance the efficacy of chemotherapeutics or targeted therapies in solid tumors, with several ongoing clinical trials.30,31,32 Inhibitors of PI3K and/or mTOR have been found in a few preclinical studies to be effective in combination with BH3 mimetics, albeit with varying rationales and outcomes and sometimes combined with a third treatment, such as radiotherapy.33,34 Most preclinical studies have combined BH3 mimetics with PI3K inhibitors, mTOR kinase inhibitors, or dual PI3K/mTOR inhibitors that are not currently approved for clinical use.35,36,37,38 It appears that combinations of BH3 mimetics with rapalogs have yet to be tested for anti-tumor activity in humans. However, preclinical studies in cell and xenograft tumor models have suggested that such combinations could be effective in some cancer settings.34,39,40,41
Here, we seek to define the effects of mTORC1 activation and inhibition on the BCL-2 family of proteins in settings with loss of the TSC tumor suppressors, with the goal of identifying strategies to enhance the therapeutic potential of mTOR inhibitors, especially rapalogs. In addition to being a tumor syndrome with a need for improved therapies, TSC has provided a powerful model for understanding the molecular, cellular, and pathological consequences of mTORC1 activation and inhibition. We find that the activation state of mTORC1 affects several pro-apoptotic and pro-survival BCL-2 family proteins in cells lacking TSC2, the most consistent of which is an mTORC1-dependent elevation in the pro-survival protein MCL-1. We find that mTORC1 inhibitors induce a potent switch in cellular dependence from MCL-1 to BCL-2 and BCL-XL for their survival. This effect can be exploited by combining rapamycin with a specific BH3 mimetic and yields a more complete and sustained response than rapamycin alone in a mouse tumor model of TSC.
Results
mTORC1 inhibitors differentially affect the expression of BCL-2 family proteins
To elucidate the effects of mTORC1 inhibitors on the core regulators of apoptosis, we utilized cells with constitutive, growth factor-independent activation of mTORC1 due to loss of the TSC2 tumor suppressor. Consistent with its known cytostatic effects, rapamycin treatment decreased the proliferation of Tsc2−/− mouse embryonic fibroblasts (MEFs) (Figure 1A). However, not only did rapamycin fail to affect cell viability, it exerted a protective effect, decreasing basal cell death, reflected by fewer cells with annexin V-positive staining (Figure 1B). Treatment of these cells or Tsc2−/− 105K renal tumor-derived cells42 with the mTORC1 inhibitors rapamycin or Torin1 resulted in various changes in transcript levels of pro-apoptotic and pro-survival BCL-2 family members. However, these effects varied between the two cell models, with only Bcl-2 transcripts being consistently upregulated by mTORC1 inhibitors in both cell lines (Figures 1C, S1A, and S1B). Immunoblot analysis of the protein products of these genes similarly revealed varied responses to a collection of distinct mTOR inhibitors, including rapamycin, the mTOR catalytic domain inhibitors Torin1 and AZD2014, and an mTORC1-selective hybrid of these two classes of inhibitors, Rapalink-1 (Figures 1D, 1E, S2A, and S2B).43,44,45,46 However, consistent changes were observed among the pro-survival BCL-2 family members, with MCL-1 being increased and BCL-2 being decreased in Tsc2-deficient cells relative to their wild-type counterparts and reversed by treatment with all 4 mTOR inhibitors (Figures 1D and 1E). Cellular BCL-XL protein levels were relatively unchanged by TSC2 status or treatment.
Figure 1.
mTORC1 inhibitors differentially affect the expression of BCL-2 family proteins
(A) The mean log2 fold change in viable cell number, via trypan blue exclusion, of Tsc2−/− MEFs treated with vehicle (DMSO) or rapamycin (20 nM) for 72h in 0.5% serum is graphed as mean ± SD relative to Day 0. n = 2. ∗p<0.05 (two-tailed Student’s t test).
(B) The percent of Tsc2−/− MEFs treated as in (A), but for 48 h. Annexin V/PI staining was quantified. n = 4.
(C) Heat maps of RT-qPCR data from Tsc2−/− MEFs and Tsc2−/− 105K renal tumor cells treated with vehicle, rapamycin (20 nM), or Torin1 (250 nM) for 24h without serum. Heat maps represent the log2 transformation of transcript levels relative to the vehicle-treated samples (graphs shown in Figures S1A and S1B). n = 2.
(D and E) Immunoblots of Tsc2+/+ and Tsc2−/− MEFs (D) and Tsc2−/− 105K cells reconstituted with TSC2 or empty vector (E) treated with vehicle (DMSO), rapamycin (20 nM), Torin1 (250 nM), AZD2014 (1 μM), or Rapalink-1 (5 nM) for 24 h without serum.
(F–I) Immunoblot of Tsc2−/− MEFs (F,H) and Tsc2−/− 105K cells (G,I) treated as above with vehicle (0h time point), rapamycin (F,G), or Torin1 (H,I) for the indicated times.
(J) Immunoblot of Rictor+/+ and Rictor−/− MEFs treated with vehicle (DMSO), or Torin1 (250 nM) for 24 h in full serum.
Although all mTOR inhibitors tested decreased MCL-1 and increased BCL-2 in these cells, the effects of the catalytic inhibitors were stronger than with rapamycin, which parallels the stronger inhibition of 4E-BP1 phosphorylation with these inhibitors, as seen by electrophoretic mobility shift (Figures 1D and 1E). A time course of rapamycin and Torin1 treatment further supports this finding, demonstrating a delayed reduction in MCL-1 levels with rapamycin relative to Torin1 treatment, whereas both inhibitors modestly increased BCL-2 levels with similar kinetics (Figures 1F–1I). Furthermore, we found that in wild-type cells with controlled levels of mTORC1 activity, MCL-1 was largely resistant to rapamycin but still sensitive to Torin1, in contrast to Tsc2−/− cells where it was sensitive to both inhibitors (Figure S2C). Given that mTOR catalytic domain inhibitors such as Torin1 inhibit both mTORC1 and mTORC2, we sought to determine whether there were contributions from mTORC2 in this regulation. Importantly, Torin1 decreased MCL-1 levels and modestly increased BCL-2 to a similar extent in Rictor+/+ and Rictor−/− MEFs, the latter missing an essential component of mTORC2 (Figure 1J). Thus, the pro-survival proteins MCL-1 and BCL-2 are reciprocally regulated by mTORC1 activation and inhibition, suggesting distinct modes of cell survival influenced by the activation state of mTORC1.
MCL-1 levels correlate with mTORC1 activation in human and mouse TSC2-deficient tumors
Because MCL-1 expression was increased in an mTORC1-dependent manner in TSC2-deficient cell lines, we next determined the status of MCL-1 in specimens of human AML, which arise because of loss of function mutations in TSC2 or TSC1.47 Histological scoring of phosphorylated ribosomal S6 (p-S6), as a marker of mTORC1 signaling, and MCL-1 staining on a tissue microarray revealed positive p-S6 staining in all 23 AML samples (14 positive, 9 strongly positive) and positive MCL-1 staining in 17 of 23 samples (13 positive, 4 strongly positive) relative to the faint staining of normal kidney tissue (Figures 2A and 2B). Because AML samples from patients treated with rapamycin are not available, we sought to determine whether tumor MCL-1 was sensitive to rapamycin using an immunocompetent syngeneic mouse TSC tumor model. This model derived from the 105K cells in C57BL/6J mice shows similar responses to rapamycin as that reported in TSC patients, with rapamycin treatment shrinking tumors, followed by regrowth on treatment withdrawal.21,22 To avoid confounding effects from tumor shrinkage, tumor-bearing mice were treated with rapamycin (1 mg/kg) or vehicle for just three consecutive days before tumor resection for immunoblot analysis. Mice treated with rapamycin displayed a decrease in the levels of tumor MCL-1, without apparent changes in BCL-2 or BCL-XL levels (Figures 2C and 2D). These data provide evidence that the effects of mTORC1 activation and inhibition on MCL-1 in our cell-based studies are also observed in tumors.
Figure 2.
MCL-1 levels correlate with mTORC1 activation in human and mouse TSC2-deficient tumors
(A–C) Representative IHC staining of the indicated human tissues from a tissue microarray. Scale bar = 0.4 mm (B) Histological scoring of p-S6 and MCL-1 staining of tumors relative to normal kidney tissue. n = 23 (C) Immunoblot of 105K xenograft tumors from mice treated with vehicle or rapamycin (1 mg/kg) for three consecutive days.
(D) Ratios of the given proteins from (C) to vinculin are graphed as mean ± SD relative to those from vehicle-treated mice. n = 4 tumors. ∗p<0.05 (two-tailed Student’s t test).
Reciprocal effects of MCL-1 and BCL-2/BCL-XL on the survival of cells upon mTORC1 inhibition
Because pro-survival BCL-2 family members block apoptosis by engaging and inhibiting pro-apoptotic activators, such as BIM, we next asked whether the differential effects of mTORC1 activation and inhibition on the cellular levels of these proteins alters the relative abundance of these protein complexes influencing cell survival. BIM immunoprecipitation revealed increased MCL-1 binding and decreased BCL-2 and BCL-XL binding in Tsc2−/− MEFs relative to Tsc2+/+ MEFs (Figure 3A). Importantly, activation of mTORC1 underlies this difference, as rapamycin treatment reversed these effects, leading to decreased BIM-MCL-1 complexes and increased BIM-BCL-2 and BIM-BCL-XL complexes relative to vehicle-treated cells. As seen above (Figure S2A), BIM levels were decreased with rapamycin in the Tsc2−/− MEFs (Figure 3A). However, normalization to BIM levels in the immune complexes confirmed that rapamycin decreases BIM engagement with MCL-1 while greatly enhancing its association with BCL-2 and BCL-XL (Figure 3B).
Figure 3.
Reciprocal effects of MCL-1 and BCL-2/BCL-XL on the survival of cells upon mTORC1 inhibition
(A) Schematic depicting BCL-2 family protein interactions to regulate apoptosis (top). Tsc2+/+ and Tsc2−/− MEFs were treated with vehicle (DMSO) or rapamycin (20 nM) for 24 h and immunoblots of proteins in BIM immunoprecipitates and lysates are shown.
(B) Ratios of the given proteins from the BIM immunoprecipitates in (A) to BIM are graphed as mean ± SD relative to those from vehicle treated wild-type cells. n = 2.
(C) The log2 fold change in viable cell number, via trypan blue exclusion, of Tsc2−/− MEFs treated with vehicle (DMSO), rapamycin (20 nM), ABT-263 (1 μM), S63845 (2 μM), or the indicated combinations for 72 h in 0.5% serum is graphed as mean ± SD relative to Day 0. n = 2.
(D and E) Cells were treated as in (C) with increasing concentrations of rapamycin and S63845 (D) or ABT-263 (E) for 48 h. Synergy scores were generated using SynergyFinder. n = 3.
(F) Tsc2−/− MEFs were pre-treated with vehicle (DMSO) or rapamycin (20 nM) for 24 h followed by continued treatment together with vehicle (DMSO) or S63845 (2 μM) for 24 h in 0.5% serum. Annexin V/PI staining was quantified. n = 2.
(G) Tsc2−/− MEFs were treated with vehicle (DMSO), rapamycin (20 nM), ABT-263 (1 μM) or the combination for 72 h in 0.5% serum. Annexin V/PI staining was quantified. n = 2. ∗p<0.05, ∗∗p<0.01 (one-way ANOVA(B) or two-way ANOVA(C)).
These findings suggest that MCL-1 is a major survival factor when mTORC1 is activated, whereas cells switch to BCL-2/BCL-XL-dependent survival on mTORC1 inhibition. To test this, we determined the effects of selective BH3-mimetic inhibitors of MCL-1 (S63845) or BCL-2 and BCL- XL (ABT-263) alone or in combination with rapamycin. Of interest, MCL-1 inhibition alone led to a dramatic loss of cells indicative of cell death, and treatment with rapamycin attenuated this effect (Figure 3C). In contrast, although both rapamycin and BCL-2/BCL-XL inhibition alone led to decreased cell numbers, only the combination induced cell death. These findings were further supported by siRNA-mediated knockdown of these proteins, with MCL-1 knockdown alone inducing cell death in a manner partially rescued with rapamycin (Figures S3A and S3B) and knockdown of BCL-2 and BCL-XL, alone or especially together, only inducing substantial cell death in the presence of rapamycin (Figures S3C and S3D). Synergy analysis was also performed using dose ranges of these inhibitors, with SynergyFinder scores above 10 reflecting synergistic interactions between the drugs and scores below −10 indicating antagonistic interactions. Consistent with our cell survival analyses, rapamycin was antagonistic to the MCL-1 inhibitor (synergy score of −17.622) but synergistic with the BCL-2/BCL-XL inhibitor (synergy score of 17.434) (Figures 3D and 3E). Measuring cell death directly, using annexin V/PI staining, confirmed that treatment with the MCL-1 inhibitor alone could kill Tsc2−/− cells and that pre-treatment with rapamycin could protect from this inhibitor (Figure 3F). Reciprocally, rapamycin enhanced the induction of cell death on treatment with the BCL-2/BCL-XL inhibitor (Figure 3G). Although not approved for clinical use and more cytotoxic than rapamycin, the structurally distinct mTOR inhibitors AZD-2014 and Rapalink-1 also strongly enhanced the induction of cell death when administered in combination with ABT-263 (Figures S3E and S3F). Importantly, this response does not depend on the p53 status of cells. Tsc2−/−p53+/+ MEFs showed similar regulation of MCL-1, BCL-2 and BCL-XL in response to mTORC1 inhibition as Tsc2−/−p53−/− MEFs, and both rapamycin and AZD-2014, likewise, enhanced the induction of cell death by ABT-263 in these p53+/+ cells (Figures S3G and S3H). Together, these data reveal that genetic activation of mTORC1 or its pharmacological inhibition can differentially influence sensitivity of cells to the emerging class of BH3 mimetics targeting pro-survival BCL-2 family proteins.
Rapamycin induces apoptosis upon inhibition of BCL-2 and BCL-XL
Given the safety and efficacy of rapalogs for the treatment of tumors in TSC and the fact that many TSC patients are already taking these drugs, we chose to further characterize the effect and therapeutic potential of combining rapamycin with BCL-2/BCL-XL inhibitors. Although treatment of Tsc2−/− tumor-derived 105K cells with rapamycin or two different doses of ABT-263 alone lead to a decrease in cell number, only the combination induced a loss of cells, indicative of cell death (Figure 4A). Importantly, rapamycin treatment fails to enhance the effects of ABT-263 in TSC2-expressing cells exhibiting normal control of mTORC1 signaling (Figures S4A and S4B). In addition, ABT-263 induced a pronounced dose-dependent increase in markers of apoptosis, cleaved caspase 3 and PARP, when co-administered with rapamycin (Figure 4B). Consistent with apoptotic cell death being induced by this combination treatment, the pan-caspase inhibitor Q-VD-OPh blocked the death of these cells (Figure 4C).
Figure 4.
Rapamycin induces apoptosis upon inhibition of BCL-2 and BCL-XL
(A) Mean log2 fold change in absorbance (570 nm) of solubilized crystal violet dye for 105K cells treated with vehicle (DMSO), rapamycin (20 nM), ABT-263 (500 nM or 1 μM), or the indicated combinations after 48 h in full serum is graphed as mean ± SD relative to Day 0. n = 2.
(B) Immunoblot of 105K cells treated with vehicle (DMSO), rapamycin (20 nM), ABT-263 (250 nM or 500 nM) or the combination for 24 h in full serum.
(C) Data from 105K cells treated with vehicle (DMSO), rapamycin (20 nM), ABT-263 (500 nM), or the indicated combinations in the presence or absence of Q-VD-OPh (20 μM) after 48 h are graphed as in (A). n = 2.
(D–G) Immunoblot of lysates from SNU-886 (D) and 97-1 (F) cells treated with vehicle (DMSO), rapamycin (20 nM), or Torin1 (250 nM) for 24 h without serum. Mean log2 fold change in absorbance (570 nm) of solubilized crystal violet dye for SNU-886 (E) and 97-1 (G) cells treated with vehicle (DMSO), rapamycin (20 nM), ABT-263 (50 nM (SNU-886), 500 nM (97-1)), or the indicated combinations after 72 h in 0.5% serum are graphed as mean ± SD relative to Day 0. n = 2.
(H) Mean log2 fold change in cell number, via trypan blue exclusion, for 105K cells 72h after transfection with the indicated siRNAs together with vehicle (DMSO) or rapamycin (20 nM) treatment in full serum is graphed as mean ± SD relative to Day 0. n = 2.
(I) Immunoblot of cells from (H).
(J–L) 105K cells were treated with vehicle (DMSO) or rapamycin (20 nM) together with increasing concentrations of ABT-199 (J), WEHI-539 (K), or ABT-263 (L) for 48 h in full serum. Viable cells, measured via CellTiter-Glo, are shown as a percentage of vehicle-treated cells and graphed as mean ± SD n = 3. ∗p<0.05, ∗∗p<0.01, ∗∗∗p<0.001, ∗∗∗∗p<0.0001 (two-way ANOVA).
Loss of function mutations in the TSC1 and TSC2 tumor suppressor genes are also found in sporadic cancers, most prevalently in liver (∼7%, predominantly TSC2) and bladder (∼13%, predominantly TSC1) (www.cbioportal.org). Thus, we characterized the response of TSC gene-deficient cell lines derived from these cancers to rapamycin for effects on pro-survival proteins and sensitivity to ABT-263 (Figures 4D–4G). MCL-1 from these human cell lines resolved as both prominent, slower migrating and apparently less abundant, faster migrating forms on immunoblots, with both species confirmed to be MCL-1 isoforms by siRNA-mediated knockdown (Figure S4C). Both rapamycin and Torin1 treatment decreased the faster migrating form of MCL-1, whereas Torin1 had a stronger effect on the slower migrating species in both the TSC2-deficient hepatocellular carcinoma line SNU-886 and the TSC1-deficient bladder cancer line 97-1 (Figures 4D and 4F). In these cells, BCL-2 is also decreased with the mTORC1 inhibitors, especially Torin1. Importantly, only the combination of rapamycin and ABT-263, and neither alone, induced the death of these cells (Figures 4E and 4G). These results expand our findings above and suggest that a rapamycin-mediated decrease in MCL-1 may be sufficient to drive cooperativity between rapamycin and ABT-263 for the induction of cell death.
We next characterized the response of cancer cells with loss of the PTEN tumor suppressor, which also exhibit growth factor-independent activation of mTORC1. In two PTEN-null prostate cancer cell lines, PC3 and LNCaP, mTORC1 inhibition decreased MCL-1 levels without substantial effects on BCL-2 or BCL-XL (Figures S4D and S4F). ABT-263 alone induced the death of these cells, and rapamycin was unable to enhance this effect (Figures S4E and S4G). Therefore, the synergistic relationship between mTOR inhibitors and BCL-2/BCL-XL inhibitors observed in mouse and human TSC-deficient cells does not appear to broadly apply to all cancer settings with genetic activation of mTORC1 signaling.
Before testing the effects of combining rapamycin with dual inhibition of BCL-2 and BCL-XL in the 105K tumor model, we determined the necessity of BCL-2 and BCL-XL inhibition, individually or together, through both genetic and pharmacological approaches. Importantly, cooperative cell death with rapamycin occurred only with siRNA-mediated knockdown of both BCL-2 and BCL-XL, revealing that these proteins play a redundant role in sustaining cell survival on rapamycin treatment (Figures 4H and 4I). Consistent with these results, the BCL-2 inhibitor ABT-199 had little effect on the viability of these cells in the presence or absence of rapamycin (Figure 4J). The BCL-XL inhibitor WEHI-539 affected cell viability, but rapamycin only modestly enhanced this effect, resulting in just a two-fold decrease in IC50 (Figure 4K). In contrast, the dual BCL-2/BCL-XL inhibitor ABT-263 greatly decreased cell viability only when combined with rapamycin, which decreased the IC50 of ABT-263 by 20-fold (Figure 4L). Together, these data confirm the necessity of inhibiting both BCL-2 and BCL-XL to induce a synergistic effect with rapamycin on cell death.
Combinatorial treatment with rapamycin and ABT-263 yields a more complete and durable anti-tumor response than rapamycin alone
To determine the in vivo relevance of the cooperative effects between mTOR inhibitors and BH3 mimetics detected in cell culture models, an immunocompetent syngeneic mouse tumor model of TSC was employed. As rapamycin exerts a strong growth-inhibitory effect in the 105K tumor model,5,21,22 our study design focused on tumor regrowth following a shorter treatment phase (Figure 5A). Treatment was initiated when tumors reached between 150 and 200 mm3, with rapamycin (1 mg/kg) or its vehicle control being administered via i.p. injection three days per week (MWF) and the BCL-2/BCL-XL inhibitor ABT-263 (100 mg/kg) or its vehicle control orally delivered daily. To capture the effects of adding BCL-2/BCL-XL inhibition to rapamycin treatment already underway, as it is in many TSC patients, ABT-263 treatment was initiated in the combination group after 7 days of rapamycin treatment. This treatment regimen was well tolerated, with no change in body weight over the course of administration (Figure S5A). At 26 days of treatment (19 days for the combination treatment), when the tumors from vehicle-treated mice reached their chosen humane endpoint (tumor volume of ∼1000 mm3), treatment was halted but with continued measurements of tumor size. During the treatment phase, ABT-263 alone slowed tumor growth, whereas both rapamycin and the combination of rapamycin and ABT-263 led to a substantial decrease in initial tumor volume (Figures 5B and 5C). The combination treatment led to a more significant decrease in tumor volume than rapamycin alone (Figures 5B, 5C, S5C, and S6B). On treatment withdrawal, the ABT-263 alone group displayed enhanced tumor growth, similar to the rate of growth in the vehicle-treated group (Figure 5B). Importantly, tumors regrew significantly faster in rapamycin-treated mice than those co-treated with ABT-263 (Figures 5B, 5D, and S6D). The rate of tumor regrowth varied between individual mice, but after 50 days of treatment withdrawal, all tumors from the rapamycin-treated group exhibited substantial regrowth, whereas only 25% (2 of 8) from the combination-treated group had surpassed their original size, with half of the tumors remaining smaller than their starting size even at 100 days post-treatment (Figure 5E). These data demonstrate a therapeutic strategy that improves the durability of rapamycin in a tumor model of TSC and prolongs survival after treatment withdrawal (Figures S5B and S5D).
Figure 5.
Combinatorial treatment with rapamycin and ABT-263 yields a more complete and durable anti-tumor response than rapamycin alone
(A) Schematic depicting experimental design.
(B) Tumor volume in mice treated with vehicle, rapamycin, ABT-263 or combinations thereof is graphed ± SEM. Rapamycin (1 mg/kg) was administered MWF (i.p.) and ABT-263 (100 mg/kg) was administered daily starting at Day 7 (oral gavage), with treatment halted at day 26. n = 6 in vehicle and ABT-263 groups, n = 8 in rapamycin and rapamycin plus ABT-263 groups.
(C) Waterfall plot of log2 fold changes in the volume of individual tumors at day 24 of treatment from (B) relative to starting tumor size (day 0).
(D) Fold change in tumor volume over the first 36 days of regrowth, following cessation of treatment, is graphed ± SEM.
(E) Tumor volume in mice from (B) plotted by individual mice.
(F) Kaplan-Meier survival curve of mice at chosen humane endpoint of tumor volume reaching 1000 mm3. ∗p<0.05, ∗∗p<0.01, ∗∗∗p<0.001, ∗∗∗∗p<0.0001 (two-way ANOVA (B), two-tailed Student’s t test (C,D), Gehan-Breslow-Wilcoxon and Mantel-Cox (F)).
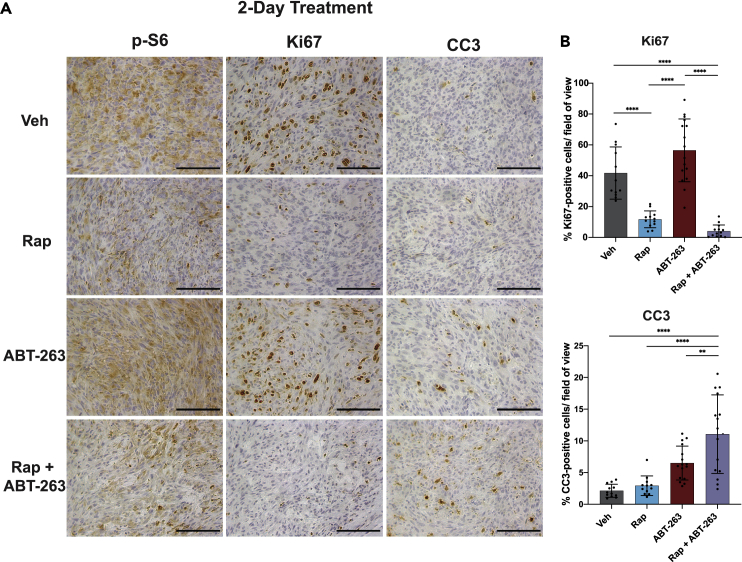
To define the nature of the improved response of tumors to the combination therapy, we compared the acute effects of the treatments above on tumor cell proliferation and apoptosis. A second cohort of mice bearing 105K xenograft tumors were treated for 1 day with rapamycin or vehicle, followed by two consecutive days of co-treatment with either ABT-263 or its vehicle control. Approximately 6 h after the final treatment, tumors were resected for IHC, which revealed that rapamycin, but not ABT-263, decreased p-S6 and Ki67 staining, indicative of a respective decrease in mTORC1 signaling and proliferation (Figures 6A and 6B). On the other hand, ABT-263, but not rapamycin, induced an increase in tumor cells with cleaved caspase 3, which was significantly enhanced when administered in combination with rapamycin. Taken together, these data extend our cell culture findings and indicate that co-treatment with ABT-263 converts rapamycin from a cytostatic to a cytotoxic therapy for improved and more sustained anti-tumor responses.
Figure 6.
Combinatorial treatment with rapamycin and ABT-263 induces apoptosis in TSC2-deficient tumors
(A) IHC analyses of tumors from mice treated once with vehicle or rapamycin (1 mg/kg, i.p.), followed by two consecutive days of the same treatment in combination with vehicle or ABT-263 (100 mg/kg, oral). Tumors were resected 6 h after the final treatment. n = 3 mice in the vehicle and rapamycin groups, n = 4 in the ABT-263 and rapamycin plus ABT-263 groups. Scale bar = 0.1 mm.
(B) Quantification of IHC sections using QuPath Image Analysis. Data are presented as mean ± SD. Sections were quantified in a blinded fashion with 3–4 non-overlapping fields counted per tumor. ∗p<0.05, ∗∗p<0.01, ∗∗∗p<0.001, ∗∗∗∗p<0.0001 (two-way ANOVA).
The translational downregulation of MCL-1 upon mTORC1 inhibition contributes to the enhanced induction of cell death by BCL-2/BCL-XL inhibitors
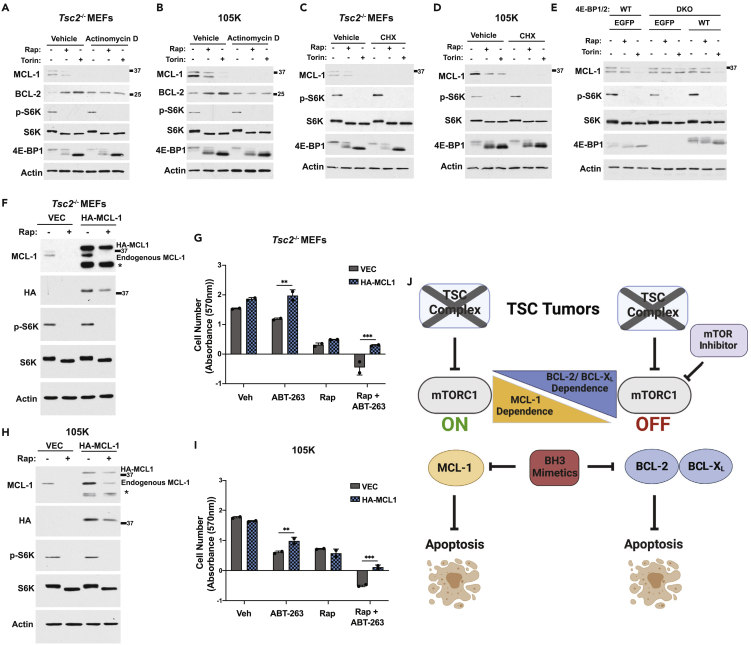
To define the mechanism underlying the effectiveness of the combination treatment of rapamycin and ABT-263 in TSC models, the molecular nature of the changes in pro-survival BCL-2 family proteins on treatment with mTOR inhibitors was determined. The MCL-1 protein has been documented to have a short half-life, whereas BCL-2 is a relatively stable protein.48 Accordingly, MCL-1 expression was lost on the inhibition of transcription with actinomycin D treatment of either Tsc2−/− MEFs or 105K cells (Figures 7A and 7B). In contrast, BCL-2 levels were unchanged, but its upregulation on rapamycin or Torin1 treatment was completely blocked. This finding is consistent with the increase in BCL-2 transcript levels observed on mTORC1 inhibition in these cells (Figures 1C, S1A, and S1B) and provides evidence that the upregulation of BCL-2 protein by mTORC1 inhibitors is likely through a transcriptional mechanism. MCL-1 protein was undetectable in Tsc2−/− MEFs and 105K cells treated for 8 h with the mRNA translation inhibitor cycloheximide, whether co-administered with mTOR inhibitors or not (Figures 7C and 7D). As suggested from previous studies,49,50,51 we hypothesized that mTORC1 signaling was influencing MCL-1 levels through its control of translation. The stronger inhibitory effects of Torin1 over rapamycin on MCL-1 protein levels further suggested a potential role for the mTORC1 substrates 4E-BP1 and 4E-BP2, the phosphorylation of which are only partially inhibited by rapamycin but completely inhibited by mTOR kinase domain inhibitors46,52(Figures 1D, 1E and 7A–7E). The 4E-BP proteins inhibit the translation initiation of mRNAs based on sequences in their 5′-untranslated regions (UTRs), whereas mTORC1 induces the translation of these mRNAs, in part, by phosphorylation and inhibition of the 4E-BPs.53 To test the involvement of the 4E-BPs in the repression of MCL-1 by mTOR inhibitors, we treated wild-type and 4ebp1/2 double knockout (DKO) MEFs with rapamycin and Torin1. As seen in other wild-type MEF lines (Figure S2C), MCL-1 was decreased with Torin1 treatment while largely insensitive to rapamycin, but MCL-1 was resistant to Torin1 in the DKO cells (Figure 7E). Importantly, the sensitivity of MCL-1 to Torin1 was restored to these cells on stable reconstitution with 4E-BP1, thus confirming that 4E-BP1 contributes to the mTORC1-mediated regulation of MCL-1 (Figure 7E).
Figure 7.
The translational downregulation of MCL-1 upon mTORC1 inhibition contributes to the enhanced induction of cell death by BCL-2/BCL-XL inhibitors
(A and B) Immunoblots of Tsc2−/− MEFs (A) and 105K cells (B) treated with vehicle (DMSO), rapamycin (20 nM), or Torin1 (250 nM) in the presence or absence of Actinomycin D (200 ng/μL) for 24 h without serum.
(C and D) Immunoblots of cells treated as in (A, B) but in the presence or absence of cyclohexamide (100 μM) for 8h.
(E) Immunoblot of wild-type (WT) or 4E-BP1/2 DKO MEFs stably expressing empty vector (EGFP) or 4E-BP1 treated with vehicle (DMSO), rapamycin (20 nM), or Torin1 (250 nM) for 24 h without serum.
(F–I) Immunoblot of Tsc2−/− MEFs (F) and 105K cells (H) expressing empty vector or HA-tagged MCL-1 lacking its 5′UTR treated with vehicle (DMSO) or rapamycin (20 nM) for 24 h without serum. ∗Apparent degradation product. Mean log2 fold change in absorbance (570 nM) of solubilized crystal violet dye for Tsc2−/− MEFs (G) and 105K cells (I) treated with vehicle (DMSO), rapamycin (20 nM), ABT-263 (500 nM (I) or 1 μM (G)), or the combination after 72 h in 0.5% serum is graphed as mean ± SD relative to Day 0. n = 2. ∗∗p<0.01, ∗∗∗p<0.001 (two-way ANOVA).
(J) Schematic summarizing findings, created with Biorender.com.
With evidence of translational regulation of MCL-1 downstream of mTORC1 in TSC models, we sought to generate a rapamycin-resistant MCL-1. We found that HA-tagged MCL-1 encoded by a cDNA lacking the MCL-1 5′-UTR was partially resistant to rapamycin when stably expressed in Tsc2−/− MEFs and 105K cells, in contrast to endogenous MCL-1 from these same cells (Figures 7F and 7H). Expression of rapamycin-resistant MCL-1 attenuated the cell death induced by the combination of rapamycin and ABT-263 in these cells (Figures 7G and 7I). These data demonstrate that the rapamycin-mediated downregulation of MCL-1 in TSC models is important to the cooperative induction of cell death by the combination of rapamycin and BCL-2/BCL-XL inhibitors.
Discussion
This study demonstrates that the activation state of mTORC1 reciprocally regulates the pro-survival BCL-2 family members, thereby differentially dictating therapeutic responses. TSC-deficient cells with constitutive activation of mTORC1 depend on MCL-1 for survival, whereas mTORC1 inhibitors in turn impose dependence on BCL-2/BCL-XL for survival (Figure 7J). Our findings reveal a therapeutic strategy that combines rapamycin, which many TSC patients are currently taking, with a dual inhibitor of BCL-2 and BCL-XL (ABT-263) that effectively converts the treatment from cytostatic to cytotoxic, thus inducing apoptosis in the context of uncontrolled mTORC1 signaling. This treatment combination has the potential to yield more complete and durable anti-tumor responses in a disease where current first-line therapies are incomplete and rapidly reversible.
The mTORC1-mediated control of MCL-1 expression is an important driver of the response to combinatorial rapamycin and ABT-263 treatment. The finding that MCL-1 expression is regulated by mTORC1 in TSC-deficient settings is consistent with preclinical studies of other cancers.35,41,49,54 However, a caveat with these previous studies is that catalytic domain inhibitors of mTORC1 and/or PI3K, and even rapamycin in some settings, also influenced the activation state of AKT, which is a major regulator of cell survival proteins, independent of mTORC1.55 The mTORC1 substrates 4E-BP1 and 4E-BP2 engage eIF4E to suppress the translation of specific mRNAs in response to mTORC1 inhibition, and we find that this suppression is required for mTOR inhibitors to decrease MCL-1. This finding is consistent with previous conclusions from studies of lymphoma models.49,50,51 In this study, the suppression of MCL-1 with mTOR inhibitors was not sufficient to induce cell death, but rather rendered cell survival dependent on BCL-2 and BCL-XL. Although the molecular mechanism is currently unknown, the upregulation of BCL-2 expression on treatment with mTORC1 inhibitors in a subset of TSC2-deficient cells is also likely to contribute to this switch in pro-survival mechanisms.
Limitations of the study
One limitation of this study is a lack of well-characterized human models of TSC, in part because of an incomplete understanding of the cell of origin of TSC-associated lesions. Although we detect increased expression of MCL-1 in human angiomyolipoma samples, a collection of samples from rapamycin-treated patients does not exist to establish its dependence on mTORC1. In addition, although we observed reproducible BCL-2 induction on mTORC1 inhibition in mouse Tsc2-deficient cell models, this effect was not seen in the tumor model or human TSC-deficient cancer cell lines. This suggests that the mTORC1 inhibitor-induced increase in BCL-2 may be context-dependent, and a reduction of MCL-1 levels is likely sufficient to drive apoptosis on co-inhibition of mTORC1 and BCL-2/BCL-XL.
There is much interest in understanding and testing the efficacy of BH3 mimetics in combination with specific chemotherapeutic agents and targeted therapies in both preclinical and clinical settings.32 The FDA-approved BCL-2 inhibitor ABT-199 (venetoclax) has emerged as a promising therapy for leukemia, both as a single agent and in combination regimens.56 The BH3 mimetic ABT-263 inhibits both BCL-2 and BCL-XL but causes on-target thrombocytopenia in vivo because of the dependence of platelets on BCL-XL.57 Although rapamycin treatment did not exacerbate this effect, a pronounced decrease in platelets was observed in ABT-263-treated mice in this current study (unpublished data). The extraordinary promise of dual BCL-2/BCL-XL inhibitors has prompted efforts dedicated to the development of novel compounds that spare platelets.58,59 Such compounds hold promise for safely targeting BCL-2 and BCL-XL in combination with rapalogs for the durable and effective treatment of tumors in TSC and other tumor syndromes, as well as in sporadic cancer, in the near future.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-4EBP1 | Cell Signaling Technology | Cat# 9644; RRID:AB_2097841 |
| Mouse monoclonal anti-alpha-tubulin | Millipore Sigma | Cat# T-5168; RRID:AB_477579 |
| Mouse monoclonal anti-beta-Actin | Millipore Sigma | Cat# A5316; RRID:AB_476743 |
| Rabbit monoclonal anti-AKT | Cell Signaling Technology | Cat# 4691; RRID:AB_915783 |
| Rabbit monoclonal anti-phospho-AKT (Thr308) | Cell Signaling Technology | Cat# 2965; RRID:AB_2255933 |
| Rabbit monoclonal anti-BAK | Cell Signaling Technology | Cat# 12105; RRID: AB_2716685 |
| Rabbit polyclonal anti-BAX | Cell Signaling Technology | Cat# 12105; RRID: AB_10695870 |
| Rabbit monoclonal anti-BCL-2 | Abcam | Cat# ab182858; RRID:AB_2715467 |
| Rabbit monoclonal anti-BCL-XL | Cell Signaling Technology | Cat# 2764; RRID: AB_2228008 |
| Rabbit polyclonal anti-BID | Bethyl | Cat# A300-084A; RRID: AB_242505 |
| Rabbit monoclonal anti-BIM | Cell Signaling Technology | Cat# 2933; RRID: AB_1030947 |
| Mouse monoclonal anti-BIM | Santa Cruz | Cat# sc-374358; RRID: AB_10987853 |
| Rabbit monoclonal anti-Cleaved Caspase-3 | Cell Signaling Technology | Cat# 9664; RRID: AB_2070042 |
| Rabbit monoclonal anti-Ki67 | Abcam | Cat# ab16667; RRID: AB_302459 |
| Rabbit monoclonal anti-MCL-1 | Cell Signaling Technology | Cat# 5453; RRID: AB_10694494 |
| Rabbit monoclonal anti-MCL-1 | Abcam | Cat# ab32087; RRID: AB_776245 |
| Rabbit polyclonal anti-PARP | Cell Signaling Technology | Cat# 9542; RRID: AB_2160739 |
| Rabbit monoclonal anti-RICTOR | Cell Signaling Technology | Cat# 9476; RRID: AB_10612959 |
| Rabbit monoclonal anti-p70 S6 Kinase | Cell Signaling Technology | Cat# 2708; RRID:AB_390722 |
| Rabbit monoclonal anti-phospho-p70 S6 Kinase (Thr389) | Cell Signaling Technology | Cat# 9234; RRID:AB_2269803 |
| Rabbit polyclonal anti-phospho-S6 (Ser235/236) | Cell Signaling Technology | Cat# 2211; RRID: AB_331679 |
| Rabbit monoclonal anti-phospho-S6 (Ser235/236) | Cell Signaling Technology | Cat# 4858; RRID: AB_916156 |
| Rabbit polyclonal anti-Vinculin | Cell Signaling Technology | Cat# 4650; RRID:AB_10559207 |
| Anti-mouse IgG, HRP-linked Antibody | Cell Signaling Technology | Cat# 7076; RRID:AB_330924 |
| Anti-rabbit IgG, HRP-linked Antibody | Cell Signaling Technology | Cat# 7074; RRID:AB_2099233 |
| Anti-rabbit IgG HRP TrueBlot | Rockland | Cat# 18-8816-31; RRID: AB_2610847 |
| Anti-normal Rabbit IgG | Cell Signaling Technology | Cat# 2729; RRID: AB_1031062 |
| Chemicals, peptides, and recombinant proteins | ||
| Rapamycin, mTORC1 inhibitor (used for in vitro studies) | Millipore Sigma | Cat# 553210; CAS: 53123-88-9 |
| Rapamycin, mTORC1 inhibitor (used for in vivo studies) | LC Laboratories | Cat# R-5000; CAS: 53123-88-9 |
| Torin1, mTOR inhibitor | Tocris | Cat# 4247; CAS: 1222998-36-8 |
| AZD2014, mTOR inhibitor | SelleckChem | Cat# S2783; CAS: 1009298-59-2 |
| RapaLink-1, mTOR inhibitor | MedChem Express | Cat# HY-111373; CAS: 1887095-82-0 |
| ABT-263 | SelleckChem | Cat# S1001; CAS: 923564-51-6 |
| ABT-199 | SelleckChem | Cat# S8048; CAS: 1257044-40-8 |
| WEHI-539 | Cayman Chemical | Cat# 21478; CAS: 1431866-33-9 |
| Q-VD-OPh | Cayman Chemical | Cat# 15260; CAS: 1135695-98-5 |
| Cycloheximide | Millipore Sigma | Cat# 01810; CAS: 66-81-9 |
| Actinomycin D | Millipore Sigma | Cat# A9415; CAS: 50-76-0 |
| Puromycin | Millipore Sigma | Cat# P8833; CAS: 58-58-2 |
| Doxycycline Hydrochloride | Millipore Sigma | Cat# D3447; CAS: 10592-13-9 |
| Crystal violet | Millipore Sigma | Cat# C6158; CAS: 548-62-9 |
| Trypan blue | Millipore Sigma | Cat #T8154; CAS:72-57-1 |
| Critical commercial assays | ||
| CellTiter-Glo Luminescent Cell Viability Assay | Promega | Cat# G7573 |
| FITC Annexin V Apoptosis Detection Kit I | BD Biosciences | Cat# 556547 |
| SuperScript III First-Strand Synthesis System | ThermoFisher Scientific/Invitrogen | Cat# 18080051 |
| Experimental models: Cell lines | ||
| Mouse: Tsc2+/+&Tsc2−/−p53−/− MEFs | Laboratory of David Kwiatkowski; Zhang et al. 2003 | N/A |
| Mouse: Tsc2−/−p53+/+ 3T3 MEFs + TSC2 | Laboratory of Brendan Manning; Huang et al. 2008 | N/A |
| Mouse: Tsc2−/−p53+/+ 3T3 MEFs + Vector | Laboratory of Brendan Manning; Huang et al. 2008 | N/A |
| Mouse: 105K | Laboratory of Lisa Henske; Filippakis et al. 2017 | N/A |
| Mouse: 105K + TSC2 | This study; Filippakis et al. 2017 | N/A |
| Mouse: 105K + Vector | This study; Filippakis et al. 2017 | N/A |
| Mouse: Rictor+/+&Rictor−/− MEFs | Laboratory of David Sabatini; Guertin et al. 2006 | N/A |
| Mouse: Eif4ebp1/2+/+&Eif4ebp1/2−/− MEFs | Laboratory of David Sabatini | N/A |
| Mouse: Eif4ebp1/2+/+ MEFs + VECTOR | This study | N/A |
| Mouse: Eif4ebp1/2−/− MEFs + VECTOR | This study | N/A |
| Mouse: Eif4ebp1/2−/− MEFs + 4E-BP1 | This study | N/A |
| Human: HEK-293T | ATCC | CRL-3216 RRID:CVCL_0063 |
| Human: 97-1 + Vector | Laboratory of David Kwiatkowski; Guo et al. 2013 | N/A |
| Human: SNU-886 | Novartis Institutes for BioMedical Research | N/A |
| Human: PC3 | ATCC | CRL-1435 RRID:CVCL_0035 |
| Human: LNCaP | ATCC | CRL-1740 RRID:CVCL_1379 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J wild-type | The Jackson Laboratory | JAX 000664 RRID:IMSR_JAX:000664 |
| Oligonucleotides | ||
| Primers for qPCR, see method details section. | IDT | N/A |
| Non-targeting control siRNA | Dharmacon/Horizon Discovery | Cat# D-001810-10-50 |
| Mcl1 siRNA | Dharmacon/Horizon Discovery | Cat# L-062229-00-0005 |
| Bcl2 siRNA | Dharmacon/Horizon Discovery | Cat# L-063933-00-0005 |
| BCL-XL (Bcl2l1) siRNA | Dharmacon/Horizon Discovery | Cat# L-065142-00-0005 |
| Recombinant DNA | ||
| psPAX2 | Addgene | 12260, RRID: Addgene_12260 |
| pMD2.G | Addgene | 12259, RRID: Addgene_12259 |
| Mouse MCL-1 (cDNA amplified from plasmid) | Addgene | 32980, RRID: Addgene_32980 |
| Mouse 4E-BP1 cDNA | This study | N/A |
| Plasmid: PLJM1-EGFP | Addgene | 19319, RRID: Addgene_19319 |
| Plasmid: PLJM1-MCL1 | This study | N/A |
| Plasmid: pTRIPZ-EGFP | Laboratory of Brendan Manning; Torrence et al. 2021 | N/A |
| Plasmid: pTRIPZ-4EBP1 | This study | N/A |
| Software and algorithms | ||
| Prism 9 | GraphPad | RRID: SCR_002798 |
| FlowJo | FlowJo | RRID: SCR_008520; https://www.flowjo.com/solutions/flowjo |
| ImageJ | Schneider et al. 2012 | RRID: SCR_003070; https://imagej.nih.gov/ij/ |
| Morpheus | Morpheus by Broad Institute | RRID: SCR_017386; https://software.broadinstitute.org/morpheus/ |
| SynergyFinder | Ianevski et al. 2020 | RRID: SCR_019318; https://synergyfinder.fimm.fi |
| QuPath | Bankhead et al. 2017 | RRID: SCR_018257; https://qupath.github.io/ |
| Other | ||
| Protein-A/G Beads Agarose Beads | ThermoFisher Scientific | Cat# 20421 |
| Matrigel Basement Membrane Matrix | BD Biosciences | Cat# 356237 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Brendan Manning (bmanning@hsph.harvard.edu).
Materials availability
Cell lines and plasmids generated in this study can be provided upon request.
Experimental model and subject details
Cell lines
Tsc2+/+p53−/− and Tsc2−/−p53−/− MEFs,60 3T3-immortalized Tsc2−/−p53+/+ MEFs stably expressing empty vector or TSC2,61Tsc2−/− 105K cells stably expressing empty vector or TSC2,42 and 97-1 cells7 were cultured in DMEM (Corning, 10-017-CV) with 10% fetal bovine serum (FBS; Gibco, 10,437-036). Rictor+/+, Rictor−/−62, Eif4ebp1/2+/+, and Eif4ebp1/2−/− MEFs were provided by D.M. Sabatini (Massachusetts Institute of Technology, Cambridge, MA) and were grown in DMEM. SNU-886 cells were provided by Novartis Institutes for BioMedical Research (Cambridge, MA) and cultured in RPMI-1640 (Corning, #10-040-CV) with 10% FBS. PC3 and LNCaP cells were purchased from ATCC. PC3 cells were grown in DMEM and LNCaP cells were grown in RPMI-1640. For experiments where media with 0.5% FBS or no FBS was used, cells were washed twice with PBS (PBS) before the addition of this media.
Animals
All animal studies were reviewed and approved by the Harvard Medical Area Standing Committee on Animals IACUC. All protocols conformed to the Guide for the Care and Use of Laboratory Animals. Xenograft tumor studies were performed in 6–7-week-old female C5BL/6J mice, which were acquired from Jackson Laboratories. The mice were group-housed (4-5 mice per cage) with water and food provided ad libitum. They were kept in a temperature controlled, pathogen-free facility with a 12:12 h light:dark cycle in standard static microisolator top cages. For all experiments, mice were randomly assigned to treatment groups such that the average starting tumor size was similar.
Method details
Chemical compounds
The following compounds were used at the specified concentrations: ABT-263 (SelleckChem #S1001 for cell culture studies; MedChem Express #HY-10087 for mouse studies), S63845 (SelleckChem, #S8383), rapamycin (EMD Millipore, #553210 for cell culture studies; LC Laboratories, #R-500 for mouse studies), Torin1 (Tocris, #4247), AZD-2014 (SelleckChem, #S2783), Rapalink-1 (MedChem Express, # HY-111373), Q-VD-OPh (Cayman Chemicals, #15260), ABT-199 (SelleckChem. #S8048), WEHI-539 (Cayman Chemicals, #21478), cycloheximide (Sigma, #01810) and actinomycin D (Sigma, #A9415).
Cell viability and death assays
Where indicated, viable cells were quantified with CellTiter-Glo Luminescent Cell Viability Assay (Promega). For synergy analysis, all samples were normalized to the untreated control, and synergy scores were calculated using the Bliss model on SynergyFinder (https://synergyfinder.fimm.fi/).63 For waterfall plots using trypan blue (Sigma, #T8154) exclusion, viable cells were counted with a hemocytometer 24 h after plating to obtain Day 0 counts. Cells were then counted 48 or 72 h after treatment. For waterfall plots using crystal violet stain, cells were washed once with PBS and stained with 0.2% crystal violet dye in 2% ethanol (Sigma, #C6158) for 10 min. After staining, cells were washed twice with water and allowed to dry. Dye was eluted with 1% SDS and absorbance was measured at 590 nm. Absorbance was measured 24 h after plating to obtain Day 0 counts and again 48 or 72 h after treatment. Annexin V/PI staining was quantified using the FITC Annexin V Apoptosis Detection Kit I (BD Biosciences, #556547). Samples were analyzed using an BD LSRFortessa Flow Cytometer and FlowJo v10.6.1 software.
qPCR
RNA was extracted using TRIzol (Invitrogen, #15596018) according to the manufacturer’s instructions. cDNA was synthesized with the Super-Script III First-Strand Synthesis System (Invitrogen, #18080051). Quantitative RT-PCR was performed on the samples using iTaq Universal SYBR Green Supermix (#1725125) on a CFX Connect Real-Time System (BioRad). Heatmaps were created using Morpheus (https://software.broadinstitute.org/morpheus/). Primer sequences: Mcl-1 (F 5′-AGAGGCTGGGATGGGTTTGT-3′, R 5′-CCCTATTGCACTCACAAGGC-3′), Bcl-2 (F 5′-GACTGAGTACCTGAACCGGC-3′, R 5′-AGTTCCACAAAGGCATCCCAG-3′), Bcl2l1 (F 5′-GCCTTTTTCTCCTTTGGCGG-3′, R 5′-TCCACAAAAGTGTCCCAGCC-3′), Bcl2l11 (F 5′-GCCAGGCCTTCAACCACTAT-3′, R 5′-TGCAAACACCCTCCTTGTGT-3′), Bid (F 5′-CCGCAAACCTTTGCCTTAGC-3′, R 5′-CAGGGAATCACCACGCAGAC-3′), Bak1 (F 5′- CCTTCTGAACAGCAGGTTGC-3′, R 5′-GACCCACCTGACCCAAGATG-3′), Bax (F 5′- AAACTGGTGCTCAAGGCCC-3′, R 5′-CTTGGATCCAGACAAGCAGC-3′), and β-actin (F 5′- CACTGTCGAGTCGCGTCC-3′, R 5′-TCATCCATGGCGAACTGGTG-3′). All samples were normalized to β-actin.
Immunoblot and immunoprecipitation
Cells were lysed in Triton X-100 buffer (40 mM HEPES pH 7.4, 120 mM NaCl, 1 mM EDTA, 1% Triton X-100, 10 mM sodium pyrophosphate, 10 mM glycerol 2-phosphate, 50 mM NaF, 0.5 mM sodium orthovanadate, 1 μM Microcystin-LR, and Sigma protease inhibitor cocktail P8340) and centrifuged at 20,000 x g for 10min at 4°C. Protein concentration was quantified via Bradford assay (Bio-Rad) and normalized between samples. For immunoprecipitations, lysates were incubated with Protein-A/G Beads Agarose Beads (ThermoFisher, #20421) and either anti-normal Rabbit IgG (CST, #2729) or anti-BIM (CST, 2933) overnight at 4°C. Beads were washed five times with Triton X-100 buffer. Input samples were 5% of lysate. Following separation by SDS-PAGE, proteins were transferred to nitrocellulose membranes, which were probed with primary antibodies overnight. Membranes were incubated with either mouse (CST, #7076) or rabbit (CST, #7074) HRP-linked secondary antibodies for 1 h and were developed using ECL Western Blotting Substrate (ThermoFisher Scientific). For detection of BCL-2 and BCL-XL in immunoprecipitates, TrueBlot Anti-Rabbit IgG HRP (Rockland, #18-8816-33) was used. ImageJ software was used for quantification.64 Membranes were probed with the following primary antibodies: MCL-1 (CST, #5453), BCL-2 (Abcam, #ab182858), BCL-XL (CST, #2764), BIM (CST, #2933), BIM (Santa Cruz sc-374358, used to probe for immunoprecipitated BIM), BID (Bethyl, #A300-084A-T), BAK (CST, #12105), BAX (CST, # 2772), phospho-S6K1-T389 (CST, #9234), S6K1 (CST, #2708), 4E-BP1 (CST, #9644), RICTOR (CST, #9476), phospho-AKT-S473 (CST, #4060), AKT (CST, #4691), cleaved caspase-3 D175 (CST, #9664), PARP (CST, #9542), vinculin (CST, #4650), and β-actin (Sigma, #A5316).
Human AML tissue study
A manual tissue-array instrument (Beecher Instruments) was utilized to construct the AML tissue microarray (TMA). The H&E-stained slides were reviewed by a pathologist for the diagnosis of renal angiomyolipoma (AML) in 21 tumor samples. Selected areas of each donor block were punched by tissue cylinders and brought into a recipient paraffin block. Each block contained normal kidney that served as control samples. A section of the TMA block was stained by H&E for histologic confirmation. IHC staining was performed on 5 μm sections of the TMA block. Sections were deparaffinized in xylene, rehydrated with graded concentrations of alcohol to distilled water, and washed in TBST. Antigen retrieval was performed in R-buffer A (Electron Microscopy Science) using a pressure cooker. 3% H2O2 was utilized to block endogenous peroxidase activity, and 5% goat serum was used to block nonspecific binding for 30 min. Slides were then incubated overnight at 4°C with the primary polyclonal rabbit anti-phospho-S6 antibody (CST, #2211) and monoclonal rabbit anti-MCL1 antibody (Abcam, #ab32087) at a dilution of 1:200 and 1:100, respectively. Slides were incubated with anti-rabbit secondary antibody (Vector Labs) at a dilution of 1:300 after being washed three times in TBST. The DakoCytomation Liquid DAB plus Substrate Chromogen System was utilized to develop peroxidase reaction. Colon adenocarcinoma tissue was used for a positive control.
Mouse studies
For tumor induction, 2.5 million 105K cells were injected subcutaneously into the flank of 6–7-week-old C5BL/6J mice (The Jackson Laboratory 000664) in a 1:1 mixture with Matrigel (BD, 356237). Tumor-bearing mice were treated as indicated in the figure legends. Tumor volume was measured every 3rd or 4th day using digital calipers.
siRNAs
Cells were transfected using Opti-MEM (Gibco, #31985062) and Lipofectamine RNAiMAX Transfection Reagent (Invitrogen, #13778150). 50 nM of the following SMARTPool ON-TARGETplus mouse siRNAs (Dharmacon/Horizon Discovery) were used: non-targeting pool (D-001810-10-50), Mcl-1 (L-062229-00-0005), Bcl-2 (L-063933-00-0005), and Bcl2l1(BCL-XL) (L-065142-00-0005).
Immunohistochemistry
Resected tumors from the TSC mouse model were fixed in 10% neutral-buffered formalin (Millipore Sigma, HT501128) for 24 h before paraffin-embedding and sectioning by the Harvard Medical School Rodent Histopathology Core, who also provided a hematoxylin and eosin (H&E) stained section. Sections were washed in Histoclear (National Diagnostics #HS-200) three times for 5 min followed by two 100% and two 95% ethanol washes for 10 min, then two washes in ddH2O for 5 min. Antigen retrieval was performed by boiling for 10 min in 10 mM sodium citrate, pH 6.0, followed by three 5-min washes in ddH2O, then 3% hydrogen peroxide for 10 min, followed by two 5-min washes in ddH2O. Sections were blocked for 1 h at room temperature in tris-buffered saline with 0.1% Tween 20 (TBST) containing 5% normal goat serum (CST, #5425). Sections were then incubated overnight at 4°C with the following primary antibodies diluted in SignalStain Antibody Diluent (CST #8112): p(S235/236)S6 (CST, #4858, 1:400), Cleaved Caspase-3 (CST, #9664 1:100), and Ki67 (Abcam, #ab16667 1:100). Slides were washed three times in TBST for 5 min and incubated in SignalStain Boost IHC Detection Reagent (CST, #8114) for 30 min at room temperature, followed by three more 5-min washes in TBST. Signal was detected using the SignalStain DAB Substrate Kit (CST, #8059) according to the manufacturer’s protocol. Sections were counterstained with hematoxylin (Sigma, #GHS132), and washed twice in ddH2O for 5 min, followed by two washes in 95% and two in 100% ethanol, then two washes in Histoclear for 10 s each. Coverslips were mounted with Permount Mounting Medium (Fisher Scientific, #SP15). Slides were imaged with an Olympus CKX41 microscope equipped with a Lumenera Infinity 2 camera. Ki67 and Cleaved Caspase 3 staining was quantified in a blinded fashion using QuPath software on 3-4 non-overlapping fields per tumor.65
cDNA constructs
cDNA encoding mouse 4E-BP1 was PCR amplified using the KOD HotStart Polymerase (EMD Millipore, #71975) and cloned into the AgeI-HF and ClaI sites of pTRIPZ-EGFP.66 Following infection and selection, transgene expression was induced with 1 μg/mL doxycycline hydrochloride (Sigma, #D3447). MCL-1 cDNA lacking its 5′UTR was amplified by PCR from the pMSCV-puro-mMcl-1 plasmid (Addgene, #32980) using primers encoding an N-terminal HA tag and cloned into the EcoRI and AgeI sites in PLJM1. For the control vector, EGFP was inserted into the PLJM1 plasmid. Lentivirus was made in HEK293T cells transfected with the plasmids pMD2.G and psPAX2 (Addgene, #12259 and #12260) and the PLJM1 constructs. After 48 h, virus-containing media was transferred to cultures of Tsc2−/− MEFs and 105K cells at a 1:1 ratio with DMEM, and cells were selected with puromycin (Sigma, #P8833). Generated plasmids were Sanger sequenced by Genewiz.
Quantification and statistical analysis
Graphical data are presented as mean ± SD or SEM as indicated in the figure legends. For all studies, n represents biological replicates, and data from all cell-based assays are representative of at least two independent experiments. An unpaired two-tailed Student’s t test was used to compare two groups. A one-way ANOVA with Tukey’s multiple comparison test was used for comparisons between three or more groups. When cells or tumors were treated with combinations of inhibitors, a two-way ANOVA with Sidak’s multiple comparison test was used. All statistical analysis was performed using GraphPad Prism 8 (GraphPad Software). For all experiments, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Acknowledgments
We thank Karen Cichowski, Taru Muranen, and Cyril Benes for critical feedback, and all members of the Manning Lab for technical assistance and discussion. This research was supported by grants from the Department of Defense’s Congressionally Directed Medical Research Program on Tuberous Sclerosis Complex, W81XWH-18-1-0370 (BDM), and the NIH: P01-CA120964 (BDM, DJK, EPH, C-LW), R35-CA197459 (BDM) and T32-ES016645 (MCM), and a kind gift from the MacPherson Fund on behalf of the Carroll family. We thank the Dana-Farber/Harvard Cancer Center for the use of the Rodent Histopathology Core (supported in part by NCI Cancer Center Support Grant NIH 5 P30 CA06516).
Author contributions
Conceptualization, M.C.M. and B.D.M.; Investigation, M.C.M., A.M.H., M.E.T., T.Z., C.F., M.W., and A.J.V.; Writing, M.C.M. and B.D.M.; Supervision, B.D.M., K.A.S., and A.J.V.; Resources, B.D.M., D.J.K., C-L.W., and K.A.S.; Funding Acquisition, B.D.M., D.J.K., E.P.H., C-LW.
Declaration of interests
B.D.M. is a member of the scientific advisory board and a shareholder of Navitor Pharmaceuticals. D.J.K. reports receiving grants from Genentech, Revolution Medicines, and AADI and consulting fees from AADI, Guidepoint, and BridgeBio Gene Therapy.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: November 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105458.
Supplemental information
Data and code availability
Original data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Ilagan E., Manning B.D. Emerging role of mTOR in the response to cancer therapeutics. Trends Cancer. 2016;2:241–251. doi: 10.1016/j.trecan.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim J., Guan K.L. mTOR as a central hub of nutrient signalling and cell growth. Nat. Cell Biol. 2019;21:63–71. doi: 10.1038/s41556-018-0205-1. [DOI] [PubMed] [Google Scholar]
- 3.Saxton R.A., Sabatini D.M. mTOR signaling in growth, metabolism, and disease. Cell. 2017;169:361–371. doi: 10.1016/j.cell.2017.03.035. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Sahra I., Manning B.D. mTORC1 signaling and the metabolic control of cell growth. Curr. Opin. Cell Biol. 2017;45:72–82. doi: 10.1016/j.ceb.2017.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henske E.P., Jozwiak S., Kingswood J.C., Sampson J.R., Thiele E.A. Tuberous sclerosis complex. Nat. Rev. Dis. Primers. 2016;2 doi: 10.1038/nrdp.2016.35. [DOI] [PubMed] [Google Scholar]
- 6.Salussolia C.L., Klonowska K., Kwiatkowski D.J., Sahin M. Genetic etiologies, diagnosis, and treatment of tuberous sclerosis complex. Annu. Rev. Genomics Hum. Genet. 2019;20:217–240. doi: 10.1146/annurev-genom-083118-015354. [DOI] [PubMed] [Google Scholar]
- 7.Guo Y., Chekaluk Y., Zhang J., Du J., Gray N.S., Wu C.L., Kwiatkowski D.J. TSC1 involvement in bladder cancer: diverse effects and therapeutic implications. J. Pathol. 2013;230:17–27. doi: 10.1002/path.4176. [DOI] [PubMed] [Google Scholar]
- 8.Ho D.W.H., Chan L.K., Chiu Y.T., Xu I.M.J., Poon R.T.P., Cheung T.T., Tang C.N., Tang V.W.L., Lo I.L.O., Lam P.W.Y., et al. TSC1/2 mutations define a molecular subset of HCC with aggressive behaviour and treatment implication. Gut. 2017;66:1496–1506. doi: 10.1136/gutjnl-2016-312734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knowles M.A., Hornigold N., Pitt E. Tuberous sclerosis complex (TSC) gene involvement in sporadic tumours. Biochem. Soc. Trans. 2003;31:597–602. doi: 10.1042/bst0310597. [DOI] [PubMed] [Google Scholar]
- 10.Chiarini F., Evangelisti C., McCubrey J.A., Martelli A.M. Current treatment strategies for inhibiting mTOR in cancer. Trends Pharmacol. Sci. 2015;36:124–135. doi: 10.1016/j.tips.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Li J., Kim S.G., Blenis J. Rapamycin: one drug, many effects. Cell Metab. 2014;19:373–379. doi: 10.1016/j.cmet.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roskoski R., Jr. Properties of FDA-approved small molecule protein kinase inhibitors: a 2022 update. Pharmacol. Res. 2022;175:106037. doi: 10.1016/j.phrs.2021.106037. [DOI] [PubMed] [Google Scholar]
- 13.Feldman M.E., Apsel B., Uotila A., Loewith R., Knight Z.A., Ruggero D., Shokat K.M. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7:e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hua H., Kong Q., Zhang H., Wang J., Luo T., Jiang Y. Targeting mTOR for cancer therapy. J. Hematol. Oncol. 2019;12:71. doi: 10.1186/s13045-019-0754-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martelli A.M., Buontempo F., McCubrey J.A. Drug discovery targeting the mTOR pathway. Clin. Sci. (Lond.) 2018;132:543–568. doi: 10.1042/CS20171158. [DOI] [PubMed] [Google Scholar]
- 16.Cabrera-Lopez C., Marti T., Catala V., Torres F., Mateu S., Ballarin J., Torra R. Assessing the effectiveness of rapamycin on angiomyolipoma in tuberous sclerosis: a two years trial. Orphanet J. Rare Dis. 2012;7:87. doi: 10.1186/1750-1172-7-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franz D.N., Leonard J., Tudor C., Chuck G., Care M., Sethuraman G., Dinopoulos A., Thomas G., Crone K.R. Rapamycin causes regression of astrocytomas in tuberous sclerosis complex. Ann. Neurol. 2006;59:490–498. doi: 10.1002/ana.20784. [DOI] [PubMed] [Google Scholar]
- 18.McCormack F.X., Inoue Y., Moss J., Singer L.G., Strange C., Nakata K., Barker A.F., Chapman J.T., Brantly M.L., Stocks J.M., et al. Efficacy and safety of sirolimus in lymphangioleiomyomatosis. N. Engl. J. Med. 2011;364:1595–1606. doi: 10.1056/nejmoa1100391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lam H.C., Nijmeh J., Henske E.P. New developments in the genetics and pathogenesis of tumours in tuberous sclerosis complex. J. Pathol. 2017;241:219–225. doi: 10.1002/path.4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bissler J.J., McCormack F.X., Young L.R., Elwing J.M., Chuck G., Leonard J.M., Schmithorst V.J., Laor T., Brody A.S., Bean J., et al. Sirolimus for angiomyolipoma in tuberous sclerosis complex or lymphangioleiomyomatosis. N. Engl. J. Med. 2008;358:140–151. doi: 10.1056/NEJMoa063564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu H.J., Lizotte P.H., Du H., Speranza M.C., Lam H.C., Vaughan S., Alesi N., Wong K.K., Freeman G.J., Sharpe A.H., et al. TSC2-deficient tumors have evidence of T cell exhaustion and respond to anti-PD-1/anti-CTLA-4 immunotherapy. JCI Insight. 2018;3 doi: 10.1172/jci.insight.98674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valvezan A.J., McNamara M.C., Miller S.K., Torrence M.E., Asara J.M., Henske E.P., Manning B.D. IMPDH inhibitors for antitumor therapy in tuberous sclerosis complex. JCI Insight. 2020;5 doi: 10.1172/jci.insight.135071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh R., Letai A., Sarosiek K. Regulation of apoptosis in health and disease: the balancing act of BCL-2 family proteins. Nat. Rev. Mol. Cell Biol. 2019;20:175–193. doi: 10.1038/s41580-018-0089-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Letai A. Apoptosis and cancer. Annu. Rev. Cancer Biol. 2017;1:275–294. doi: 10.1146/annurev-cancerbio-050216-121933. [DOI] [Google Scholar]
- 25.Sarosiek K.A., Letai A. Directly targeting the mitochondrial pathway of apoptosis for cancer therapy using BH3 mimetics - recent successes, current challenges and future promise. FEBS J. 2016;283:3523–3533. doi: 10.1111/febs.13714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Billard C. BH3 mimetics: status of the field and new developments. Mol. Cancer Ther. 2013;12:1691–1700. doi: 10.1158/1535-7163.MCT-13-0058. [DOI] [PubMed] [Google Scholar]
- 27.Del Gaizo Moore V., Brown J.R., Certo M., Love T.M., Novina C.D., Letai A. Chronic lymphocytic leukemia requires BCL2 to sequester prodeath BIM, explaining sensitivity to BCL2 antagonist ABT-737. J. Clin. Investig. 2007;117:112–121. doi: 10.1172/JCI28281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng J., Carlson N., Takeyama K., Dal Cin P., Shipp M., Letai A. BH3 profiling identifies three distinct classes of apoptotic blocks to predict response to ABT-737 and conventional chemotherapeutic agents. Cancer Cell. 2007;12:171–185. doi: 10.1016/j.ccr.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 29.Konopleva M., Contractor R., Tsao T., Samudio I., Ruvolo P.P., Kitada S., Deng X., Zhai D., Shi Y.-X., Sneed T., et al. Mechanisms of apoptosis sensitivity and resistance to the BH3 mimetic ABT-737 in acute myeloid leukemia. Cancer Cell. 2006;10:375–388. doi: 10.1016/j.ccr.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 30.Cragg M.S., Harris C., Strasser A., Scott C.L. Unleashing the power of inhibitors of oncogenic kinases through BH3 mimetics. Nat. Rev. Cancer. 2009;9:321–326. doi: 10.1038/nrc2615. [DOI] [PubMed] [Google Scholar]
- 31.Merino D., Kelly G.L., Lessene G., Wei A.H., Roberts A.W., Strasser A. BH3-Mimetic drugs: blazing the trail for new cancer medicines. Cancer Cell. 2018;34:879–891. doi: 10.1016/j.ccell.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 32.Townsend P.A., Kozhevnikova M.V., Cexus O.N.F., Zamyatnin A.A., Jr., Soond S.M. BH3-mimetics: recent developments in cancer therapy. J. Exp. Clin. Cancer Res. 2021;40:355. doi: 10.1186/s13046-021-02157-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fulda S. Synthetic lethality by co-targeting mitochondrial apoptosis and PI3K/Akt/mTOR signaling. Mitochondrion. 2014;19:85–87. doi: 10.1016/j.mito.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 34.Kim K.W., Moretti L., Mitchell L.R., Jung D.K., Lu B. Combined Bcl-2/mammalian target of rapamycin inhibition leads to enhanced radiosensitization via induction of apoptosis and autophagy in non-small cell lung tumor xenograft model. Clin. Cancer Res. 2009;15:6096–6105. doi: 10.1158/1078-0432.CCR-09-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Faber A.C., Farago A.F., Costa C., Dastur A., Gomez-Caraballo M., Robbins R., Wagner B.L., Rideout W.M., 3rd, Jakubik C.T., Ham J., et al. Assessment of ABT-263 activity across a cancer cell line collection leads to a potent combination therapy for small-cell lung cancer. Proc. Natl. Acad. Sci. USA. 2015;112:E1288–E1296. doi: 10.1073/pnas.1411848112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H., Liu L., Chang H., Zou Z., Xing D. Downregulation of MCL-1 and upregulation of PUMA using mTOR inhibitors enhance antitumor efficacy of BH3 mimetics in triple-negative breast cancer. Cell Death Dis. 2018;9:137. doi: 10.1038/s41419-017-0169-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pareja F., Macleod D., Shu C., Crary J.F., Canoll P.D., Ross A.H., Siegelin M.D. PI3K and Bcl-2 inhibition primes glioblastoma cells to apoptosis through downregulation of Mcl-1 and Phospho-BAD. Mol. Cancer Res. 2014;12:987–1001. doi: 10.1158/1541-7786.MCR-13-0650. [DOI] [PubMed] [Google Scholar]
- 38.Preuss E., Hugle M., Reimann R., Schlecht M., Fulda S. Pan-mammalian target of rapamycin (mTOR) inhibitor AZD8055 primes rhabdomyosarcoma cells for ABT-737-induced apoptosis by down-regulating Mcl-1 protein. J. Biol. Chem. 2013;288:35287–35296. doi: 10.1074/jbc.M113.495986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ackler S., Xiao Y., Mitten M.J., Foster K., Oleksijew A., Refici M., Schlessinger S., Wang B., Chemburkar S.R., Bauch J., et al. ABT-263 and rapamycin act cooperatively to kill lymphoma cells in vitro and in vivo. Mol. Cancer Ther. 2008;7:3265–3274. doi: 10.1158/1535-7163.MCT-08-0268. [DOI] [PubMed] [Google Scholar]
- 40.Gardner E.E., Connis N., Poirier J.T., Cope L., Dobromilskaya I., Gallia G.L., Rudin C.M., Hann C.L. Rapamycin rescues ABT-737 efficacy in small cell lung cancer. Cancer Res. 2014;74:2846–2856. doi: 10.1158/0008-5472.CAN-13-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iacovelli S., Ricciardi M.R., Allegretti M., Mirabilii S., Licchetta R., Bergamo P., Rinaldo C., Zeuner A., Foa R., Milella M., et al. Co-targeting of Bcl-2 and mTOR pathway triggers synergistic apoptosis in BH3 mimetics resistant acute lymphoblastic leukemia. Oncotarget. 2015;6:32089–32103. doi: 10.18632/oncotarget.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Filippakis H., Alesi N., Ogorek B., Nijmeh J., Khabibullin D., Gutierrez C., Valvezan A.J., Cunningham J., Priolo C., Henske E.P. Lysosomal regulation of cholesterol homeostasis in tuberous sclerosis complex is mediated via NPC1 and LDL-R. Oncotarget. 2017;8:38099–38112. doi: 10.18632/oncotarget.17485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chung J., Kuo C.J., Crabtree G.R., Blenis J. Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 protein kinases. Cell. 1992;69:1227–1236. doi: 10.1016/0092-8674(92)90643-q. [DOI] [PubMed] [Google Scholar]
- 44.Guichard S.M., Curwen J., Bihani T., D'Cruz C.M., Yates J.W., Grondine M., Howard Z., Davies B.R., Bigley G., Klinowska T., et al. AZD2014, an inhibitor of mTORC1 and mTORC2, is highly effective in ER+ breast cancer when administered using intermittent or continuous schedules. Mol. Cancer Ther. 2015;14:2508–2518. doi: 10.1158/1535-7163.MCT-15-0365. [DOI] [PubMed] [Google Scholar]
- 45.Rodrik-Outmezguine V.S., Okaniwa M., Yao Z., Novotny C.J., McWhirter C., Banaji A., Won H., Wong W., Berger M., de Stanchina E., et al. Overcoming mTOR resistance mutations with a new-generation mTOR inhibitor. Nature. 2016;534:272–276. doi: 10.1038/nature17963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thoreen C.C., Kang S.A., Chang J.W., Liu Q., Zhang J., Gao Y., Reichling L.J., Sim T., Sabatini D.M., Gray N.S. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giannikou K., Malinowska I.A., Pugh T.J., Yan R., Tseng Y.Y., Oh C., Kim J., Tyburczy M.E., Chekaluk Y., Liu Y., et al. Whole exome sequencing identifies TSC1/TSC2 biallelic loss as the primary and sufficient driver event for renal angiomyolipoma development. PLoS Genet. 2016;12:e1006242. doi: 10.1371/journal.pgen.1006242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adams K.W., Cooper G.M. Rapid turnover of mcl-1 couples translation to cell survival and apoptosis. J. Biol. Chem. 2007;282:6192–6200. doi: 10.1074/jbc.M610643200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bi C., Zhang X., Lu T., Zhang X., Wang X., Meng B., Zhang H., Wang P., Vose J.M., Chan W.C., et al. Inhibition of 4EBP phosphorylation mediates the cytotoxic effect of mechanistic target of rapamycin kinase inhibitors in aggressive B-cell lymphomas. Haematologica. 2017;102:755–764. doi: 10.3324/haematol.2016.159160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mills J.R., Hippo Y., Robert F., Chen S.M., Malina A., Lin C.J., Trojahn U., Wendel H.G., Charest A., Bronson R.T., et al. mTORC1 promotes survival through translational control of Mcl-1. Proc. Natl. Acad. Sci. USA. 2008;105:10853–10858. doi: 10.1073/pnas.0804821105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wendel H.G., Silva R.L., Malina A., Mills J.R., Zhu H., Ueda T., Watanabe-Fukunaga R., Fukunaga R., Teruya-Feldstein J., Pelletier J., et al. Dissecting eIF4E action in tumorigenesis. Genes Dev. 2007;21:3232–3237. doi: 10.1101/gad.1604407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choo A.Y., Yoon S.O., Kim S.G., Roux P.P., Blenis J. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc. Natl. Acad. Sci. USA. 2008;105:17414–17419. doi: 10.1073/pnas.0809136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Richter J.D., Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- 54.Jebahi A., Villedieu M., Petigny-Lechartier C., Brotin E., Louis M.H., Abeilard E., Giffard F., Guercio M., Briand M., Gauduchon P., et al. PI3K/mTOR dual inhibitor NVP-BEZ235 decreases Mcl-1 expression and sensitizes ovarian carcinoma cells to Bcl-xL-targeting strategies, provided that Bim expression is induced. Cancer Lett. 2014;348:38–49. doi: 10.1016/j.canlet.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 55.Fulda S. Shifting the balance of mitochondrial apoptosis: therapeutic perspectives. Front. Oncol. 2012;2:121. doi: 10.3389/fonc.2012.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Juarez-Salcedo L.M., Desai V., Dalia S. Venetoclax: evidence to date and clinical potential. Drugs Context. 2019;8:212574. doi: 10.7573/dic.212574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gandhi L., Camidge D.R., Ribeiro de Oliveira M., Bonomi P., Gandara D., Khaira D., Hann C.L., McKeegan E.M., Litvinovich E., Hemken P.M., et al. Phase I study of Navitoclax (ABT-263), a novel Bcl-2 family inhibitor, in patients with small-cell lung cancer and other solid tumors. J. Clin. Oncol. 2011;29:909–916. doi: 10.1200/JCO.2010.31.6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Khan S., Zhang X., Lv D., Zhang Q., He Y., Zhang P., Liu X., Thummuri D., Yuan Y., Wiegand J.S., et al. A selective BCL-XL PROTAC degrader achieves safe and potent antitumor activity. Nat. Med. 2019;25:1938–1947. doi: 10.1038/s41591-019-0668-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lv D., Pal P., Liu X., Jia Y., Thummuri D., Zhang P., Hu W., Pei J., Zhang Q., Zhou S., et al. Development of a BCL-xL and BCL-2 dual degrader with improved anti-leukemic activity. Nat. Commun. 2021;12:6896. doi: 10.1038/s41467-021-27210-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang H., Cicchetti G., Onda H., Koon H.B., Asrican K., Bajraszewski N., Vazquez F., Carpenter C.L., Kwiatkowski D.J. Loss of Tsc1/Tsc2 activates mTOR and disrupts PI3K-Akt signaling through downregulation of PDGFR. J. Clin. Investig. 2003;112:1223–1233. doi: 10.1172/jci17222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang J., Dibble C.C., Matsuzaki M., Manning B.D. The TSC1-TSC2 complex is required for proper activation of mTOR complex 2. Mol. Cell Biol. 2008;28:4104–4115. doi: 10.1128/MCB.00289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guertin D.A., Stevens D.M., Thoreen C.C., Burds A.A., Kalaany N.Y., Moffat J., Brown M., Fitzgerald K.J., Sabatini D.M. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev. Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 63.Ianevski A., He L., Aittokallio T., Tang J. SynergyFinder: a web application for analyzing drug combination dose-response matrix data. Bioinformatics. 2020;36:2645. doi: 10.1093/bioinformatics/btaa102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bankhead P. Developing image analysis methods for digital pathology. J. Pathol. 2022;257:391–402. doi: 10.1002/path.5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Torrence M.E., MacArthur M.R., Hosios A.M., Valvezan A.J., Asara J.M., Mitchell J.R., Manning B.D. The mTORC1-mediated activation of ATF4 promotes protein and glutathione synthesis downstream of growth signals. Elife. 2021;10 doi: 10.7554/eLife.63326. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.