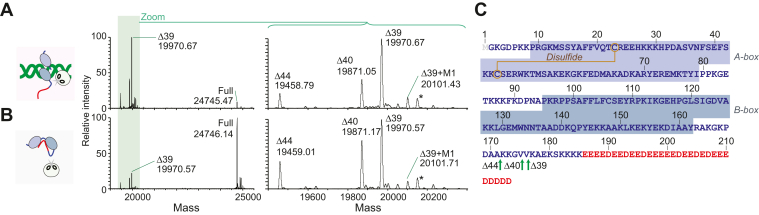
Figure 2.
LC-MS intact mass-based identification of the cleavage sites for the DNA-mediated proteolysis of disulfide HMGB1 by neutrophil elastase.A and B, deconvolved mass spectra produced for the reaction mixtures in the presence (panel A) and absence (panel B) of 20 bp DNA. The regions for the reaction products and the original disulfide HMGB1 protein are shown. In the presence of DNA, Δ39 was the major cleavage product and minor cleavage products were Δ40 and Δ44. The peaks indicated by “Δ39 + M1” correspond to the Δ39 product of the HMGB1 protein retaining the initial methionine M1. The peaks indicated by asterisks correspond to the Δ39 product of the gluconoylated HMGB1 lacking M1. The minor species with M1 or gluconoyl modification (73) were also observed for mass spectra recorded for the original proteins (see Fig. S4 in the Supporting information). For more detailed information about the species identified by LC-MS, see Table S1 in the Supporting information. C, amino acid sequence of HMGB1 and the identified cleavage sites (indicated by the green arrows).