Figure 2.
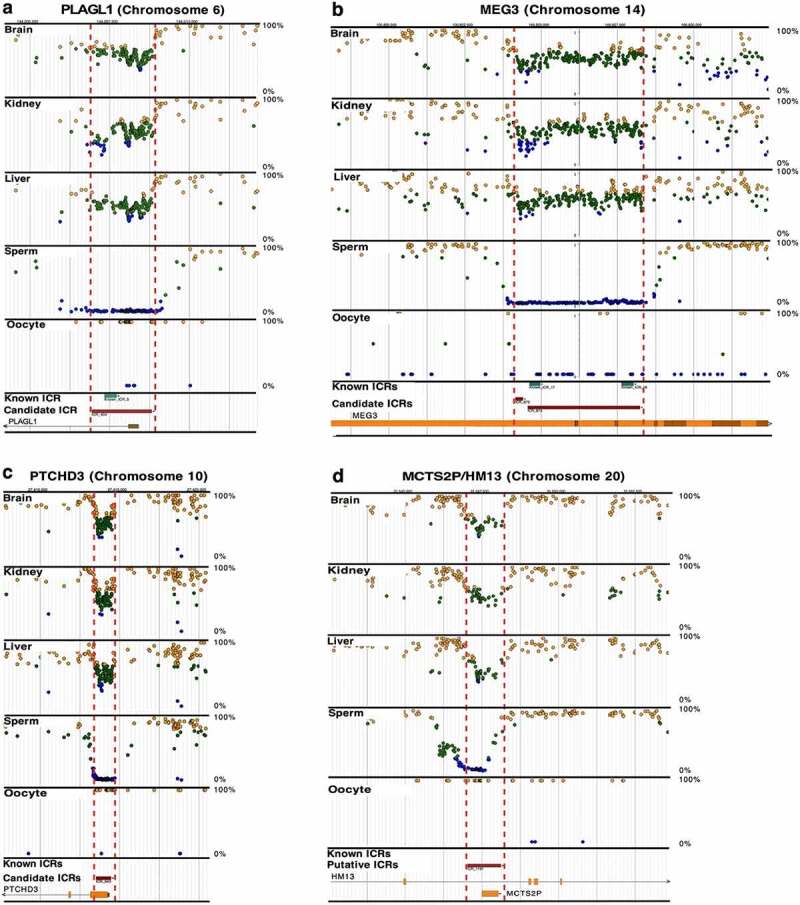
Previously known and candidate ICRs. (a) PLAGL1 (ICR_404) and (b) MEG3 (ICR_872,873) imprinted genes contain previously known ICRs (blue boxes) [14]. They were also identified in our whole-genome bisulphite sequencing (WGBS) screen for candidate ICRs (red boxes); visualize at https://humanicr.org/. The ICRs defined herein overlap and extend beyond the currently known ICRs. (c) PTCHD3 (ICR_643) and (d) MCTS2P/HM13 (ICR_1191) contain only candidate ICRs (red boxes) determined in this study, and potentially control novel imprinted genes. The methylation levels for ICRs is 50 ± 15% for the candidate ICRs. DNA methylation levels determined by WGBS in sperm and oocytes are also provided. These sequence data were supplemented with publicly available gametic sequence (accession number JGAS00000000006) and part of the control sperm data from PRJNA754049 [25,26]. Dots indicate hemi-methylation (green), hypomethylation (blue), and hypermethylation (yellow) based on WGBS. The vertical dashed red lines delineate the candidate ICR regions.
