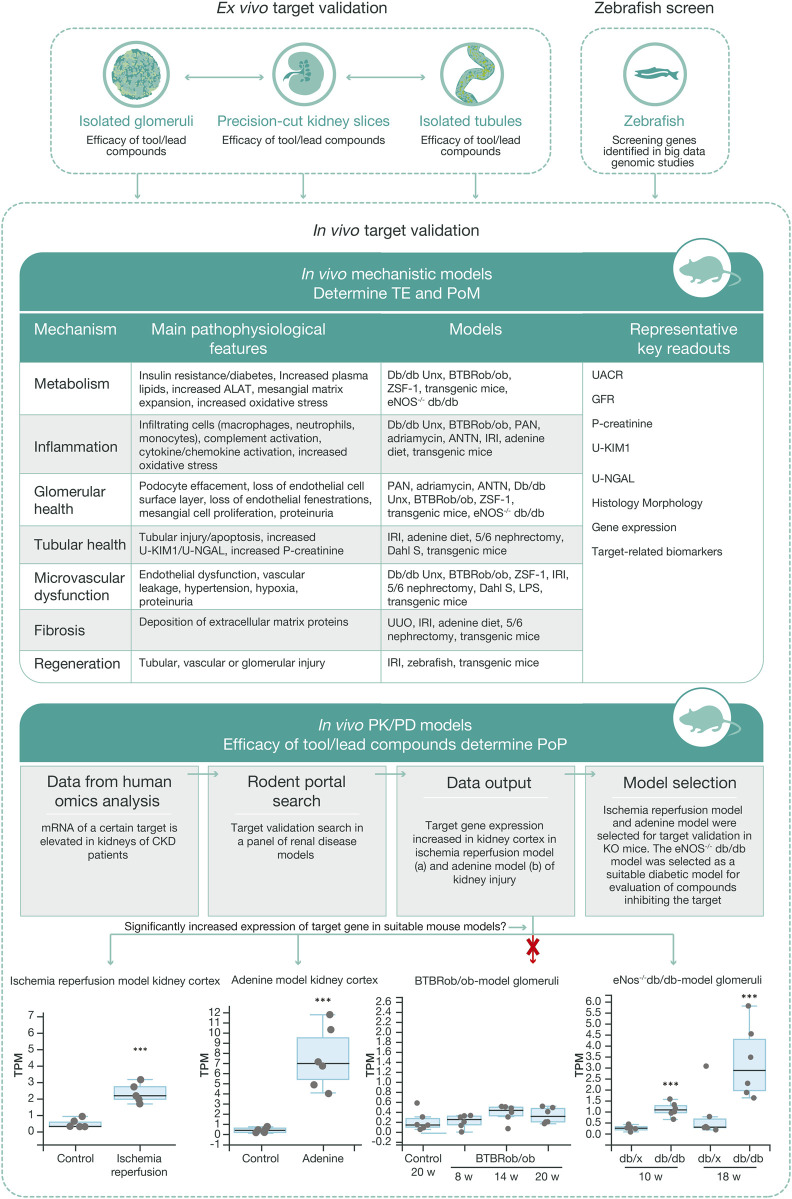
FIGURE 5.
Workflow for in vivo target validation and compound testing. Ex vivo models are chosen to validate genes of interest and assess the efficacy of compounds targeting these gene products, based on their cellular expression and target engagement, respectively. Data from the ex vivo experiments guide the design of mechanistic and PKPD in vivo studies that generate data regarding target engagement and proof of mechanism, and on exposure and proof of principle, respectively. Various mechanistic models are used to validate targets and measure efficacy of test compounds. Selection of the most relevant in vivo PKPD model for a specific target or signaling pathway is based on ex vivo experimental data, human omics data, and RNA sequencing data from our ‘rodent portal’, generated from a panel of rodent renal disease models. The bottom panel illustrates differential upregulation of a target gene in different disease models, which in turn allows selection of the most appropriate model for the tested protocol. ALAT, alanine aminotransferase; ANTN, accelerated/non-accelerated nephrotoxic nephritis; BTBR, black and tan brachyuric; eNOS, endothelial nitric oxide synthase; IRI, ischemic reperfusion injury; KIM1, kidney injury molecule one; LPS, lipopolysaccharide; NGAL, neutrophil gelatinase; PAN, pyromycin aminonucleoside; PKPD, pharmacokinetic–pharmacodynamic; POM, proof of mechanism; TE, target engagement; UUO, unilateral ureter obstruction.