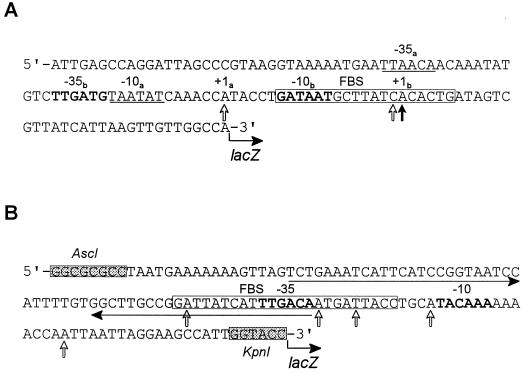
FIG. 3.
Nucleotide sequences of the p1 (A) and p2 (B) promoter regions. (A) The 123-bp fragment containing the p1 promoter (upstream of hmuP′) was cloned into a single-copy-number lacZ reporter plasmid, pEU730, generating pHMU44. (B) The 130-bp fragment of the intergenic sequence between hmuR and hmuS containing the p2 promoter region was cloned into pEU730, generating pHMU55. The shaded boxes shown in the p2 sequence are AscI and KpnI restriction sites that were designed into the oligonucleotide primers used in PCR. Potential FBS (in open boxes) were identified within p1 and p2; the p2 FBS is 5 bp longer than the E. coli consensus sequence (GATAATGATAATCATTATC). The solid vertical arrow in the p1 promoter region indicates the probable transcriptional start site of the major primer extension product of RNA isolated from Y. pestis KIM6+ or KIM6-2044.1(pHMU4)+. Open vertical arrows in both the p1 and p2 sequences show potential transcriptional start sites of the minor primer extension products of RNA isolated from KIM6+ (p1 only) and KIM6-2044.1(pHMU4)+. Boldface letters or underlined sequences indicate potential −35 and −10 regions. Within the p1 sequence, the −35b and −10b regions correspond to the major transcriptional start site (+1b), and the −35a and −10a regions may correspond to a weaker transcriptional start site (+1a). The horizontal arrows show imperfect inverted repeats within the p2 promoter region.