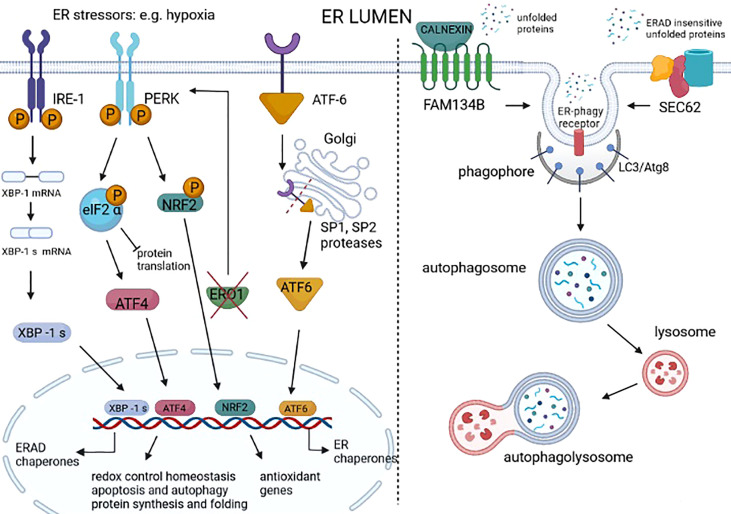
Figure 2.
Unfolded protein response (UPR) and ER-phagy in cancer. UPR is a homeostatic response to ER stress that is present in many cancer types. UPR is activated by three different sensors on the ER membrane: IRE1, PERK, and ATF6. IRE1 dimerizes following ER stress, activating an RNAse domain that promotes the unconventional splicing of XBP1 mRNA (XBP1s). The translated XBP1s acts as a transcription factor of genes involved in ER-associated degradation (ERAD) and chaperones. ER stress–activated PERK phosphorylates eIF2 alpha, promoting the attenuation of protein translation while also promoting the phosphorylation of NRF2, thereby the transcription of genes with an antioxidant function. The PERK signal also favors the selective translation of ATF4, which regulates the redox control, the genes involved in autophagy, and the CHOP-ERO1 axis. Regarding the axis CHOP-ERO1, we have seen that in breast cancer cells under hypoxic conditions, ERO1 is not downstream to CHOP. However, the lack of ERO1 converges and activates the PERK signal (21). ATF6 translocates in the Golgi where it is cleaved by SP1 and SP2 proteases and acts as a transcription factor of ER chaperones. ER stress also activates ER-phagy, a mechanism that leads to the clearance of ER portions containing misfolded proteins. FAM134B is an ER-phagy receptor that, through a physical interaction with a protein adaptor, such as calnexin, might sense unfolded proteins and starts the autophagy of the ER. SEC62 is another ER-phagy receptor that leads to the ER-phagy of ER portions containing ERAD-insensitive unfolded proteins.