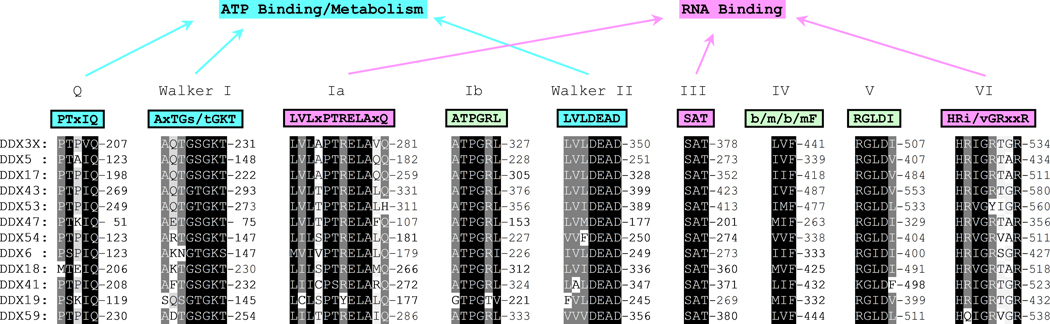
Fig 1. Illustration of the nine highly conserved protein motifs within the Rec-A like domains of DEAD box RNA helicases.
Protein sequences of DDX3, DDX5, DDX17, DDX43, DDX53, DDX47, DDX54, DDX6, DDX18, DDX41, DDX19, and DDX59 were obtained from the NCBI database as Fasta formats. In all cases of multiple isoforms, isoform 1 was used. Alignment was done in ClustalX2 with default settings (Gap opening: 10, Gap Extension: 0.2, and protein weight matrix: Gonnet series) and shading in GENEDOC. In all cases the sequences are represented by single letter amino acid code. The conserved consensus motif sequences above their respective shaded alignments are given with abbreviations: x = any amino acid and b = branched chain amino acids. Designations of each motif (Q, Walker I, Ia, Ib, Walker II, III, IV, V, & VI) are above the consensus sequences and those involved in ATP binding/metabolism and RNA binding are indicated by arrows.