Figure 5.
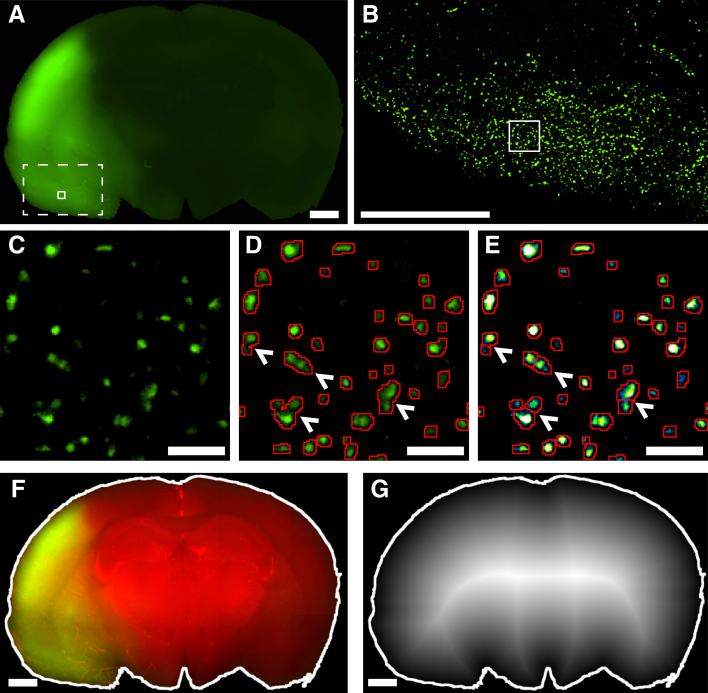
Image processing allows for the segmentation of individual leukocytes and the mapping of their exact position across the infarct. A: The raw green fluorescent protein (GFP) channel showing the inherent autofluorescence of the infarct compared with healthy tissue. The dashed box denotes the area shown in B, and the solid box shows the area illustrated in C–E. B: Image after preprocessing for the region marked by the dashed box in A indicating removal of nonuniform background enabling visualization of cells. C: Zoomed in region of the solid box in A and B of the preprocessed image. D: Thresholding alone is ineffective in resolving overlapping cells because the region between cells is above the threshold used for segmentation. The arrowheads indicate cells that are not effectively delineated from each other. E: The image in D represents using a heatmap to help visualize local maxima. The Laplacian of Gaussian filter highlights the local maxima, which were used as markers for the marker-based watershed transform for separating overlapping cells. The arrowheads indicate cells from D that have become effectively delineated from each other after the application of the Laplacian of Gaussian filter and watershed transform. F: The brain boundary highlighted in white was determined using both the GFP and vessel channel (shown in red, for better contrast). G: Distance transform of the entire brain boundary used to calculate the distance of a given point from the surface, with higher (whiter) intensity denoting greater distance. (Note that this has been done for the entire brain, not just the infarcted side.) The cell centroid position was mapped onto this and used to determine the distance of each cell from the nearest cortical surface. Scale bars: 1 mm (A, B, F, and G); 50 μm (C–E).