Fig. 1.
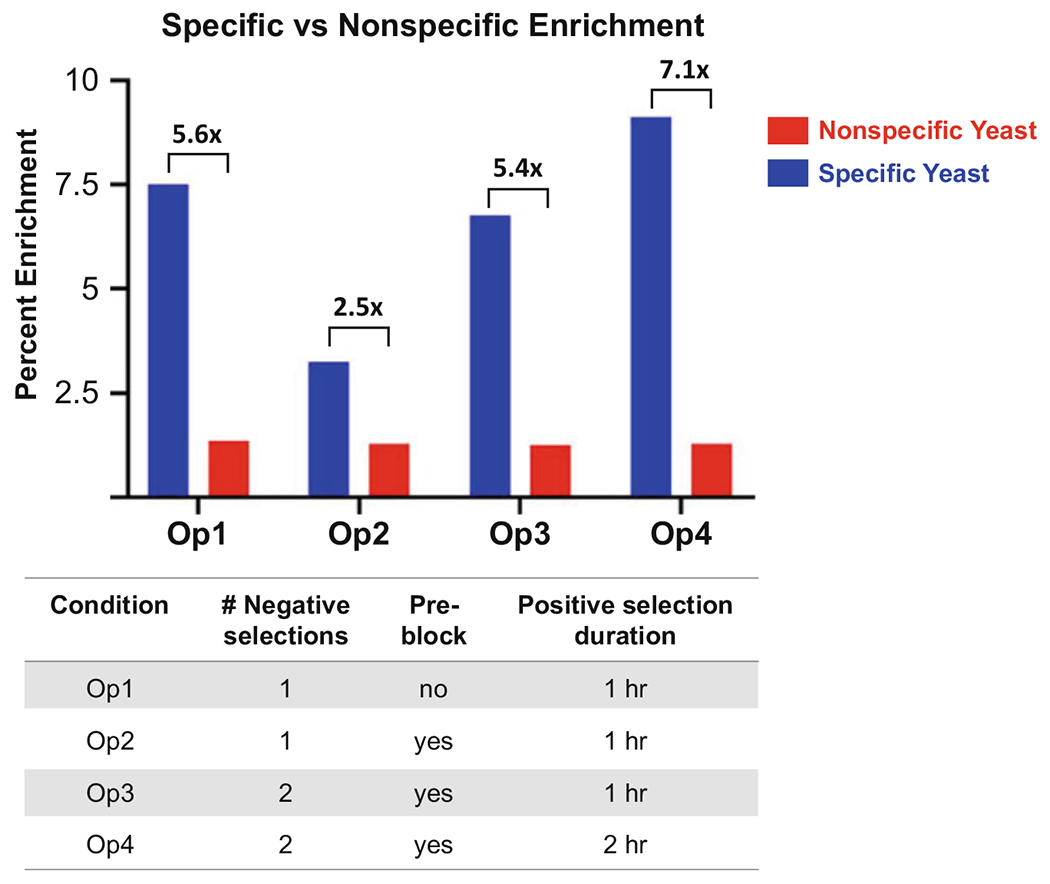
Enrichment of specific (blue) versus nonspecific (red) scFv-expressing yeast following one round of adherent cell-based selections under four different selection conditions. An anti-programmed death-ligand 1 (PD-L1) scFv-expressing yeast clone was spiked into a naïve library of 109 diversity [16], and 1010 total yeast were subjected to a single round of biopanning using the following conditions: (i) One 30 min negative selection (NS) + 60 min positive selection (PS); (ii) One 30 min NS + 30 min pre-block with soluble target-null mammalian cells at a 2:1 target-null:target-expressing cell ratio (PB) + 60 min PS; (iii) Two 30 min NS + 30 min PB + 60 min PS; and (iv) Two 30 min NS + 30 min PB + 120 min PS. Results showed that the additional NS and implementation of the PB did not significantly impact specific enrichment ratios, and in fact, implementation of PB with only a single NS actually decreased specific enrichment. However, lengthened PS incubation time (120 min) in combination with an additional NS and PB resulted in the highest specific enrichment ratio
