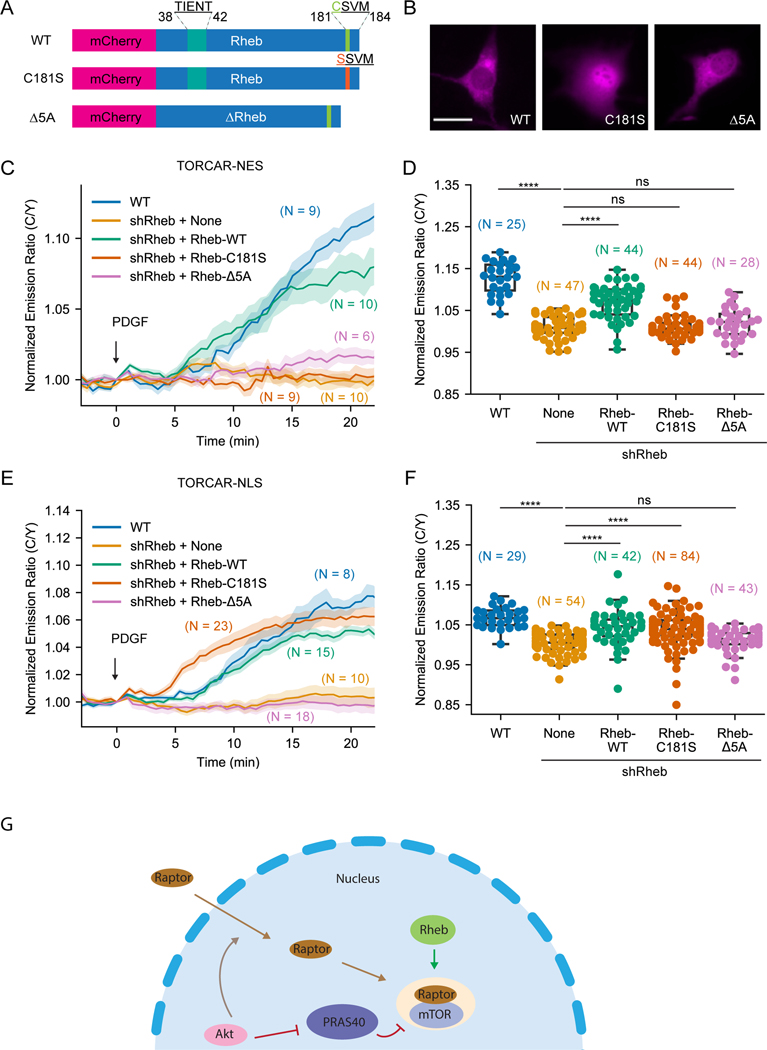
Figure 4. Farnesylation-deficient Rheb had different effects on rescuing subcellular mTORC1 activity.
(A) Schematics of different Rheb constructs tagged with mCherry. (B) Representative images of overexpressed mCherry-Rheb constructs in NIH3T3 cells from 3 independent experiments. (C-D) Representative averaged time course traces (C) and summary (D) of PDGF-induced maximum responses of TORCAR-NES in double starved NIH3T3 cells. Blue, wild type cells (WT, N = 25); yellow, Rheb knockdown cells (shRheb + None, N = 47 cells); green, Rheb knockdown cells transfected with wild type mCherry-Rheb-WT (shRheb + Rheb-WT, N = 44); orange, Rheb knockdown cells transfected with mCherry-Rheb-C181S (shRheb + Rheb-C181S, N = 44); pink, Rheb knockdown cells transfected with mCherry-Rheb-Δ5A (shRheb + Rheb-Δ5A, N = 28 cells). (E-F) Representative averaged time course traces (E) and summary (F) of PDGF-induced maximum responses of TORCAR-NLS in double starved NIH3T3 cells. Blue, wild type cells (WT, N = 29); yellow, Rheb knockdown cells (shRheb + None, N = 54); green, Rheb knockdown cells transfected with wild type mCherry-Rheb-WT (shRheb + Rheb-WT, N = 42); orange, Rheb knockdown cells transfected with mCherry-Rheb-C181S (shRheb + Rheb-C181S, N = 84); pink, Rheb knockdown cells transfected with mCherry-Rheb-Δ5A (shRheb + Rheb-Δ5A, N = 43). (G) Model showing that active Rheb in the nucleus is critical for activating nuclear mTORC1, in addition to the previously reported Akt-mediated nuclear translocation of Raptor and phosphorylation of PRAS40. N indicates the cell number quantified from at least 3 independent experiments. Shaded areas indicate standard error of the mean (SEM). Box plots show the upper and lower adjacent values, interquartile range and the median. Scale bar = 10 µm. See also Figure S3.