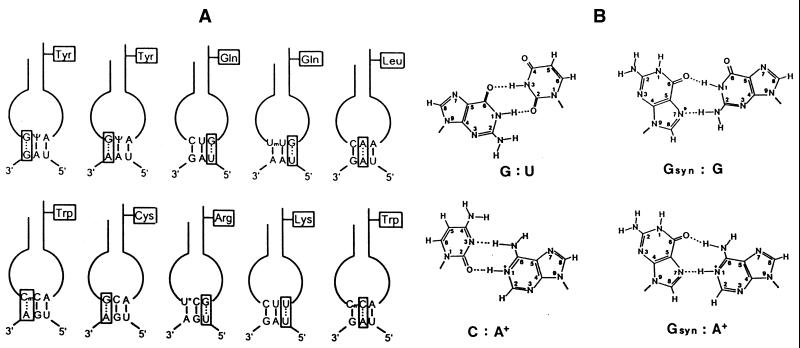
Figure 12.
Unconventional base interactions involved in the misreading of termination codons by natural suppressor tRNAs. (A) Schematic presentation of putative anticodon–codon interactions by eukaryotic suppressor tRNAs. With the exception of tRNALys(CUU) and tRNATrp(CmCA) misreading the UAG codon, all types of non-canonical codon reading by means of an appropriate suppressor tRNA have been demonstrated directly in vitro (19,39,48,52,54,63,70,89). In the case of the two former examples, incorporation of lysine and tryptophan (in addition to tyrosine) has been elucidated by amino acid sequence analysis of the translation protein produced by readthrough of an in-frame UAG codon within the yeast Ste6 gene (78). The tRNAGln isoacceptors with a UmUG anticodon from Tetrahymena, Nicotiana and mouse liver are able to also read UAG besides UAA (41,46,48). (B) Hypothetical interactions between non-complementary bases. In purine–purine mismatches, the nucleoside of the anticodon is presented in the syn configuration (isomerisation about the glycosyl bond of the nucleotide). The space-filling cyclopentene-diol side chain of the queuine base is attached to guanine at the indicated position (*) via a C–C bond, thus impairing the proposed G–G interaction (39). The adenosine in G:A and C:A mismatches is shown in the protonated form.