Abstract
Increasing preclinical and clinical results have demonstrated that mRNA vaccines efficiently prevent infectious diseases and are safe in animal models and humans. In this study, we fabricated a multivalent influenza mRNA lipid nanoparticle (LNP) vaccine with mRNAs of hemagglutinins from influenza H1N1 and H3N2 viruses, matrix protein 1, and nucleoprotein. We found that cutaneous immunization with mRNA LNPs induced strong Th1 and Th2 cellular immunity with robust antigen-specific antibody titers and increased cytokine-secreting splenocytes and antibody-secreting cells. The supplement of cGAMP improved the immunogenicity of mRNA LNPs. Compared with αGC or cGAMP/αGC adjuvanted mRNA LNP formulations in our study, cGAMP mRNA LNPs induced more robust antibody responses. Enhanced cellular immunity with more IL-4 and IFN-γ secreting cells and effector memory T cell populations in spleens, as well as increased CD4+ resident memory (TRM) T cells in lungs were observed in cGAMP mRNA LNPs immunized group. These results demonstrated that cGAMP is an effective adjuvant for cutaneous vaccination of multivalent mRNA LNP vaccines in mice to induce stronger immune responses in the spleen and lung, and the cGAMP-adjuvanted mRNA LNPs protected against homologous and heterologous viral infection.
Keywords: MT: Delivery Strategies, mRNA lipid nanoparticle, influenza virus, cutaneous vaccination, adjuvant, cGAMP
Graphical abstract

Adjuvant encapsulated multivalent mRNA lipid nanoparticle vaccine induces strong immune responses conferring cross-protections in mice.
Introduction
Influenza viral infection has caused severe global health concerns annually since the outbreak of the 1918 influenza pandemic. Vaccination has been recognized as an effective countermeasure against viral influenza infection. Vaccines aid in public health by reducing the severity of illness, lowering the hospitalization rate, and protecting people with certain chronic health conditions. Conventional seasonal flu vaccines are trivalent or quadrivalent vaccines containing influenza A and B viruses predicted to circulate in the coming flu season.
Influenza viral antigens continuously evolve by acquiring genetic mutations, which results in circulating viruses diverging from the predicted strains, reducing the vaccines’ protective efficiency. The influenza vaccine is typically less effective against influenza A/H3N2 viruses because viruses of this type acquire more frequent antigenic mutations than influenza A/H1N1 and influenza B viruses.1,2 Egg-based vaccine production can also reduce a vaccine’s effectiveness by inducing egg-related adaptations in the virus stock. Thus, a more advanced technique is needed to improve the protective efficacy of influenza vaccines. Compared with time-consuming egg- or cell-based vaccine production, mRNA production can be rapidly modified to a new viral sequence without unpredictable viral adaptations when an emerging pandemic has been confirmed.
In recent years, mRNA technology and nanotechnology combinations have been successfully applied to novel vaccine strategies. Preclinical and clinical results have demonstrated that mRNA vaccines could work efficiently to prevent infectious diseases and cancers and are safe and well tolerated in animal models and humans.3,4 Various nanoplatforms have been studied for mRNA packaging and delivery to improve the immunogenicity of mRNA vaccines.5 Of them, mRNA lipid nanoparticles (LNPs) have proven safe and effective. Beyond packaging and delivering the mRNA, the empty LNP or liposomes can exhibit intrinsic adjuvanticity and enhance the efficacy of vaccines.6,7,8 The SARS-CoV-2 mRNA lipid nanoparticle vaccines are examples of mRNA LNPs recently approved for human use in the United States by the FDA.9,10 In recent studies, influenza mRNA LNP vaccines have increased humoral and cellular immune responses and protected against influenza virus challenges.11,12,13,14
Beyond new technologies for formulating vaccines, advanced novel adjuvants can also be effective tools for improving vaccine efficacy. The utilization of adjuvants to improve vaccine immune responses has been widely studied. Alum, MF59, AS01, and CpG 1018 have been used in human vaccines in the United States. Codelivery of appropriate adjuvants with mRNA vaccines is an effective strategy to enhance the immune response. For example, adjuvanted mRNA vaccine nanoparticles improved the effectiveness of cancer immunotherapy.15,16
In this study, we examined the application of the adjuvants 2′3′- cyclic GMP-AMP (cGAMP) and α-GalCer (αGC). cGAMP is a natural STING agonist with a negative charge and has been demonstrated as an effective adjuvant for influenza vaccines.17,18 αGC is a potential mucosal adjuvant capable of inducing mucosal secretory IgA (sIgA) and systemic IgG responses against viral infections.19,20,21 Nanoplatforms incorporating αGC activated iNKT cells and generated augmented antigen-specific CTL responses.22,23,24,25
Besides biocompatible packaging, delivery, and adjuvant-based enhancement, for an influenza vaccine to be effective, it must induce immune responses to targets found on or within the virus. Seasonal flu vaccines elicit antibody responses to the rapidly evolving viral surface antigen hemagglutinin (HA). Compared with influenza surface antigens, internal influenza proteins, such as matrix protein 1 (M1) and the nucleoprotein (NP), are highly conserved antigens with the potential to induce broadly reactive T cell immunity.26,27,28 Substantial M1- and NP-specific T cell responses have increased protection against influenza infection.27,29,30 Heterologous protection has been granted by T cell immune responses like lung resident memory CD8 T cells.31,32 Thus, our mRNA formulation will include mRNA of HA antigens (HA1 and HA3) from both HA phylogenic groups targeting robust antibody responses and M1 and NP aiming at broadly reactive specific T cell responses.
In this study, we developed novel influenza mRNA vaccines by incorporating mRNAs of multiple influenza antigens with cGAMP and/or αGC into LNPs. Mice were cutaneously immunized with the resulting mRNA LNP vaccines. We found that cGAMP-adjuvanted multivalent mRNA vaccines triggered strong cellular immune responses and provided protection against diverse influenza infections.
Results
Fabrication and characterization of influenza mRNA lipid nanoparticles
We used in vitro transcription to generate codon-optimized mRNAs encoding the open reading frames of influenza HAs (PR8 HA1 and Aichi HA3), PR8 M1, PR8 NP, and non-translational regions (diagrammed in Figure S1A). Transcribed mRNAs were purified, and the expression of the resulting mRNAs was determined at 12 h and 24 h post-transfection in 293T cells (Figures S1B and S1C).
Figure 1A depicts a schematic diagram of mRNA LNPs containing the lipid mixture, mRNAs, and adjuvants. The size of individual HA1, HA3, M1, and NP LNPs ranged from 80 to 142 nm by dynamic light scattering analysis (Figures S2A and S2B). LNPs showed zeta potentials of +30 to +40 mV (Figure S2D). The concentrations of total and unencapsulated mRNAs were measured by the RiboGreen RNA reagent, indicating an over 90% encapsulation efficiency (EE%) (Figure S2C).
Figure 1.
Fabrication and characterization of influenza mRNA lipid nanoparticles
(A) A diagram of adjuvanted influenza mRNA lipid nanoparticles (LNPs). (B and C) The sizes and zeta potential values of mRNA encapsulated LNPs, cGAMP-adjuvanted, and αGC/cGAMP double-adjuvanted mRNA LNPs. (D) Transmission electron microscopy of mRNA LNPs. Scale bars represent 0.5 μm. (E and F) The stimulation of IFN-β and IFN-γ protomer activities in 293T cells after being transfected with different doses of particulate cGAMP or soluble cGAMPs. (G) The formation of αGC:CD1d complex on BMDCs after being stimulated with particulate αGC or soluble αGCs (s- αGCs). (H), (I), and (J) The cytokine production from BMDCs after LNPs and cGAMP and/or αGCs adjuvanted LNPs stimulation. The histograms were presented as mean ± SEM. Statistical significance was analyzed by F-test and two-tailed t test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; n.s. represented no significance. (E)–(J) show the results of one experiment that is representative of two independent experiments.
293T cells were transfected with these LNPs to determine the protein expression and analyzed by western blots (Figure S2E). Because of their different protein expression patterns, HA3 and M1 expressions were detected at 12 h after transfection, while HA1 and NP expressions were analyzed 24 h after transfection to observe optimal expressions. Immune fluorescence assays showed the membrane localization of HA1 and HA3 and the cytoplasmic distribution of the internal proteins (NP and M1) (Figure S3). These results indicated we successfully generated different mRNA constructs from influenza antigens and encapsulated the mRNAs into LNPs.
Characterization of cGAMP and αGC-adjuvanted mRNA LNPs
To improve the immunogenicity of our mRNA vaccines, we further generated mRNA LNPs with cGAMP and αGC co-encapsulated for comparison. The sizes and zeta potentials of the resulting multivalent influenza mRNA LNPs are shown in Figures 1B and 1C. The LNPs showed similar size distribution that peaked at 100 nm. Their zeta potentials were between +30 and +40 mV. The morphology of these LNPs was nearly spherical by transmission electron microscopy (Figure 1D).
To better understand the effectiveness and the immune correlates of protection of our vaccine candidates, we determined the induction of downstream IFN-β and IFN-γ in the STING signaling pathway. 293T cells overexpressing STING and IFN-β or IFN-γ promoter plasmids were transfected with different concentrations of soluble cGAMP (s-cGAMP) or cGAMP lipid nanoparticles (p-cGAMP). Compared with unstimulated cells (control) or 1 μg of s-cGAMP-transfected cells, the transfection with 2 μg of s-cGAMP significantly increased the IFN-β-mediated signal. Furthermore, 1 μg of p-cGAMP induced three times more IFN-β luciferase activity than 2 μg of s-cGAMP (Figure 1E). We used 1 μg of cGAMP per 10 μg of mRNA in our LNPs for later animal studies. However, neither the soluble cGAMP nor the cGAMP encapsulated particles stimulated IFN-γ signaling activity (Figure 1F).
CD1d is a constitutively expressed cell surface protein on antigen-presenting cells (APCs), such as dendritic cells (DCs). APCs present CD1d-loaded αGC for recognition by invariant natural killer T (iNKT) cells. The activation of iNKT cells results in the production of Th1 and Th2 cell-associated cytokines.23,33 To understand the role of our vaccine candidates in this signaling pathway, we determined the percentages of αGC:CD1d-complex-positive DCs by using bone-marrow-derived dendritic cells (BMDCs) isolated and cultured from the mice (Figure 1G). We stimulated the BMDCs with 2.5% of αGC-presenting lipid particles containing 1 μg of cGAMP (2.5% p-αGC/cGAMP) and 1% or 5% of soluble αGC (1% s-αGC and 5% s-αGC) separately. In comparison, p-cGAMP and unstimulated cells (control) were included as controls. Figure 1G shows that 14.4% and 38.1% of CD11c+ DCs were loaded with αGC after incubation with 1% and 5% of soluble αGC, respectively. We detected significantly increased αGC:CD1d+CD11c+ populations (61.9%) when the BMDCs were stimulated with αGC in the particulate forms. These results indicated that the nanoparticle-displayed αGC could improve the loading efficiency on CD1d, which might induce stronger iNKT activation.
The secretion of different cytokines from DCs at the early immunization phase is important to modulate the types of T cell responses.34 To determine the effect of different LNP formulations on DCs, we stimulated BMDCs with mRNA LNPs with and without cGAMP and/or αGC (designated as LNPs, cGAMP-LNPs, αGC-LNPs, and cGAMP/αGC-LNPs). Unstimulated BMDCs (mock) were included as controls. IFN-α and IFN-β are well-defined type I IFNs that affect innate and adaptive cellular immune responses through managing DC maturation and migration, promoting T cell responses, and enhancing B cell responses.35
As shown in Figures 1H and 1I, cGAMP-adjuvanted mRNA LNPs (cGAMP-LNPs) induced significantly increased expression of IFN-α and IFN-β. In contrast, LNPs alone or αGC adjuvanted mRNA LNP (αGC-LNPs) stimulation could not induce the secretion of IFN-α and IFN-β. cGAMP and αGC double-adjuvanted LNPs promoted high levels of both IFN-α and IFN-β. DCs are potent producers of IL-12, a dominant cytokine in Th1 responses.36,37 Different LNP formulations induced increased IL-12 expression (Figure 1J). These results indicate that encapsulating cGAMP in mRNA LNPs improves innate immune responses.
Humoral immune responses and protection induced by influenza mRNA LNPs
To evaluate the immunogenicity of mRNA LNPs, Balb/c mice were immunized via intradermal injection twice with the mRNA LNP vaccines with or without cGAMP and αGC, and the Aichi virus-specific antibodies were analyzed 3 weeks after boost. For the multivalent mRNA LNP groups, each dose consisted of 5 μg of mRNA for each influenza antigen of Aichi HA3, PR8 HA1, NP, and M1 (designated as Mix-LNPs). Meanwhile, the Aichi-LNP formulation contained 10 μg of Aichi HA3 mRNA as a control. The cGAMP, αGC, or cGAMP/αGC adjuvanted Mix-LNPs were named cGAMP-LNPs, αGC-LNPs, and cGAMP/αGC-LNPs.
Immune sera from Aichi-LNPs, Mix-LNPs, and cGAMP-LNPs groups showed comparable Aichi-specific IgG, IgG1, and IgG2a at different dilutions (Figures 2A–2C). Compared with sera from cGAMP-LNP-immunized mice, IgG antibody levels were significantly lower in the cGAMP/αGC double-adjuvanted LNPs group at the 1:900 dilution (Figure 2A). Although there were no significant IgG1 level differences among different groups (Figure 2B), reduced IgG2a was observed in the αGC-LNPs group compared with the cGAMP-LNPs group (Figure 2C).
Figure 2.
Humoral immune responses induced by various influenza mRNA lipid nanoparticles
Mice sera were collected at 3 weeks after boosting immunization and used for analysis. (A, B, and C) The optical density (OD) values at 450 nm for different serum dilutions of Aichi-specific IgG (A), IgG1 (B), and IgG2a (C) were determined by ELISA. (D) The mouse body weight loss and (E) survival rate after Aichi viral challenge. Serum IgG specific to (F) A/Aichi/2/1986 HA3, (G) A/New York/55/2004 HA3, (H) A/mallard/Alberta/455/2015 HA4, (I) A/Puerto Rico/8/1934 HA1, and (J) A/California/04/2009 HA1 by ELISA, respectively. The histograms were presented as mean ± SEM. Statistical significance was analyzed by F-test and two-tailed t test. ∗p < 0.05; ∗∗p < 0.01. Data represented one experiment with n = 4 mice per group.
At week 4 after boosting immunization, the mice were challenged with 15x LD50 of A/Aichi/2/1968 (A/Aichi), and their body weights were measured daily for 14 days. The naive mouse body weights fell below 80% within 7 days after the high-lethal dose of viral challenge. However, the mice immunized with Aichi-LNPs, Mix-LNPs, and cGAMP-LNPs had no symptoms and kept their initial body weights during the 14 days (Figure 2D). While various LNP-immunized groups completely survived, significant body weight loss was observed in the αGC-LNPs and cGAMP/αGC-LNPs groups by day 5 after infection in contrast to the cGAMP-LNPs group (Figures 2D and 2E).
The reactivities to various influenza viral HAs from two phylogenetic groups (Group 1 and Group 2)were evaluated by ELISA. Immune sera from Mix-LNP-immunized and other adjuvanted LNP-immunized mice showed strong reactivities to HA3 and HA4 from Group 2 viruses (Figures 2F–2H), as well as the homologous HA1 from Group 1 (Figure 2I). Weak cross-reactivity to the rHA of the 2009 pandemic H1N1 was observed (Figure 2J). These data demonstrated that immunization of multivalent mRNA LNPs could induce strong antibody responses with lower doses of dominant antigen mRNA, and encapsulation of cGAMP into these LNPs maintained the ability to stimulate robust humoral immune responses. In contrast, αGC incorporation into LNPs impaired the serum IgG2a titers and protection against homologous viral infection. The levels of IgG2a to IgG1 indicated that LNP immunization induced balanced Th1 and Th2 immune responses.
Cellular immune responses induced by multivalent influenza mRNA LNPs
Based on the previous analysis of humoral immunity, we focused our studies on cGAMP-adjuvanted multivalent mRNA LNPs and further evaluated the cellular immune responses 4 weeks after the boosting immunization. The multivalent mRNA LNPs without an adjuvant (LNPs) or cGAMP/αGC double-adjuvanted LNPs (cGAMP/αGC-LNPs) were included as controls. The influenza mRNA LNPs induced significantly increased Aichi virus-, PR8 virus-, NP-, and M1-specific IgG antibody titers 3 weeks after the boosting immunization (Figure 3A), as well as higher Aichi and PR8 viral-specific IgG1 and IgG2a antibody titers (Figures 3B and 3C).
Figure 3.
Cellular immune responses
(A) Aichi-, PR8-, NP-, and M1-specific IgG endpoint titers in mouse immune sera. (B and C) Aichi- and PR8-specific IgG2a and IgG1 endpoint titers. (D and E) Aichi, PR8, and NP peptide-specific IL-4- and IFN-γ-secreting splenocytes. (F) Aichi- and PR8-specific IgG-secreting cells in spleens. (G and H) NP147-155-specific CD8 T cell populations. (I) Aichi- and PR8-specific IgG-secreting plasma cells in the bone marrow. The histograms were presented as mean ± SEM. Statistical significance was analyzed by F-test and two-tailed t test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; n.s. represented no significance. (A)–(C) show the results of one experiment with the sera from eight mice in each group. (D)–(I) show the results of one experiment with three independent replicates for each sample. Mice groups were n = 5.
Compared with the naive group, mRNA LNP immunization demonstrated stronger cellular immune responses with significantly increased numbers of IL-4 and IFN-γ-secreting splenocytes after Aichi and PR8 viral stimulation by ELISpot assays (Figures 3D and 3E). As shown in Figure 3D, mice in the cGAMP-LNPs group produced significantly increased Aichi- and PR8-specific IL-4-secreting splenocytes compared with the LNPs and cGAMP/αGC-LNPs groups. Splenocytes from cGAMP-LNPs-immunized mice showed more robust IFN-γ secretion after PR8 stimulation (Figure 3E).
Most conserved influenza CD8 T cell epitopes during influenza infections are derived from NP. NP-specific CD8+ cytotoxic T cells (CTLs) are important for viral clearance and recovery from influenza infection. We used NP peptides as stimulators to evaluate the NP-specific cellular immune responses by flow cytometry. As shown in Figures 3D and 3E, all LNP-immunized groups showed increased numbers of NP-specific IL-4 and IFN-γ-secreting splenocytes, among which cGAMP-LNPs presented the highest numbers of NP-specific IL-4 and IFN-γ-secreting cells. Furthermore, we observed significantly increased NP tetramer and CD8 double-positive populations in splenocytes from mRNA LNP-immunized mice (Figures 3G and 3H).
We also determined the antigen-specific antibody-secreting cells (ASCs) in mouse spleens and bone marrow by ELISpot. Multivalent LNP immunization significantly enhanced the numbers of Aichi- and PR8-specific ASCs in spleens (Figure 3F) and bone marrow (Figure 3I). The cGAMP-LNPs group exhibited elevated ASCs compared with double-adjuvanted LNP immunization (Figures 3F and 3I). These results demonstrated that the adjuvanticity of cGAMP improved the mRNA LNP-induced cellular immunity, while the αGC and cGAMP combination attenuated the cellular immune responses.
Protective efficacy against homologous and heterologous influenza virus challenges
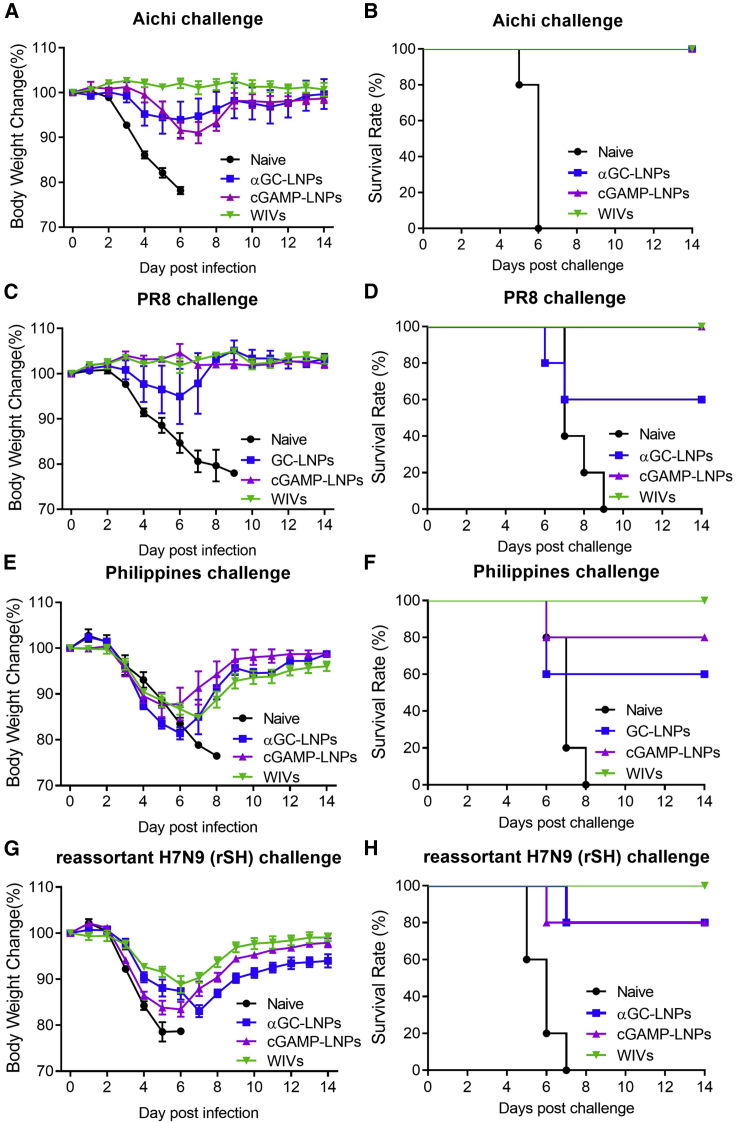
To investigate the protective efficacy of the cGAMP-LNP vaccines, mice were given homologous viral challenges of 5x LD50 of A/PR8/1934 (A/PR8) and 30x LD50 of A/Aichi, and heterologous viral challenge of 3x LD50 of A/Philippines/2/1982 (A/Phi) and reassortant H7N9 (rSH) viruses 4 weeks after the cGAMP-LNP-boosting immunization. Groups of mice were immunized with whole inactivated virus vaccines (WIVs) containing 1 μg each of A/Aichi and A/PR8 WIVs or αGC adjuvanted multivalent mRNA LNPs (αGC-LNPs) as controls.
The cGAMP-LNPs, αGC-LNPs, and WIVs immunizations conferred complete protection against A/Aichi viral infection (Figures 4A and 4B). However, the αGC-LNP immunization showed a lower survival rate (60%) against the A/PR8 homologous viral challenge than the cGAMP-LNPs or WIVs group (Figures 4C and 4D). Mice who received the cGAMP-LNPs, αGC-LNPs, or WIVs had 10%–15% body weight loss during the A/Phi (Figure 4E) or rSH (Figure 4G) infection. The WIVs group showed a 100% survival rate against the A/Phi or rSH challenge, while the cGAMP-LNP immunization conferred partial protection with an 80% survival rate (Figures 4F and 4H). The αGC-adjuvanted LNPs produced an even lower survival rate (60%) against A/Phi (Figure 4F).
Figure 4.
Protective efficacy against homologous and heterologous influenza virus challenge
Mice were challenged with 30xLD50 of A/Aichi, 5xLD50 of A/PR8, 3xLD50 of A/Phi, or 3xLD50 of rSH, and the mouse body weights and survivals were monitored for 14 days. (A and B) A/Aichi; (C and D) A/PR8; (E and F) A/Phi; (G and H) rSH. Data were presented as mean ± SEM. Mice groups were n = 5.
We evaluated antibody levels from immune sera 3 weeks after boosting immunization. Like the WIVs immunization group, cGAMP-adjuvanted multivalent mRNA LNPs induced high Aichi-specific IgG titers (Figure S4A). WIVs induced a Th1-biased immune response with significantly higher IgG2a levels, while the cGAMP-LNPs immunization showed more balanced Th1 and Th2 immune responses, indicating less proinflammatory responses. These results suggested that the cGAMP-adjuvanted influenza mRNA LNP vaccines provided complete protection against homologous strain challenges and partial protection against heterologous viral infection.
Lung histological examinations were performed 6 days after the A/Aichi challenge. The naive group showed severe inflammation with intense tissue damage and leukocyte infiltration. By contrast, mRNA LNP-immunized mice presented less immune cell infiltration and inflammation (Figure 5A). Consistent with the histological results, different LNP vaccine immunized groups had significantly reduced lung viral titers (Figure 5B).
Figure 5.
The antiviral responses in the pulmonary area after mRNA lipid nanoparticles vaccination
The prime and boost vaccination mice were infected with 5XLD50 of Aichi. Different groups of mice were euthanized on day 6 after infection, and the lungs and washes were collected for analysis. (A) Histology analysis of lungs by H&E staining. Scale bars represent 200 μm. (B) Lung viral titers. (C) Aichi-specific IgG levels in BALF. (D and E) The percentages of NP147-155-specific CD8 T cell populations in the pulmonary lymphocytes. (F and G) The CD4 and CD8 T cell population in BALF lymphocytes. (H, I, and J) The IL-12/p40, TNF-α, and IL-4 levels in BALFs. The histograms were presented as mean ± SEM. Statistical significance was analyzed by F-test and two-tailed t test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; n.s. represented no significance. Data represented one experiment with n = 5 mice per group.
Furthermore, lung lymphocytes were isolated, and NP tetramer-specific CD8+ T cell populations were analyzed. After virus infection, we detected significantly increased NP-tetramer+CD8+ T cell populations in the lungs (Figures 5D and 5E) and significantly enhanced CD4+ and CD8+ T cell populations in bronchoalveolar lavage fluids (BALF) from cGAMP-LNP-immunized mice (Figures 5F, 5G, and S5). The highest level of Aichi-specific IgG was detected in BALF samples of the cGAMP-LNPs group (Figure 5C).
We measured the expression of TNF-α, IL-12/p40, and IL-4 in BALF. Figures 5H–5J show that naive mice produced high IL-12/p40 and TNF-α after virus infection. In contrast, the mice in the LNPs, cGAMP-LNPs, or αGC/cGAMP-LNPs groups displayed significantly reduced IL-12/p40 and TNF-α levels (Figures 5H and 5I). Significantly enhanced IL-4 was detected in BALF from the mRNA LNPs-immunized mice (Figure 5J). These results indicated that LNP intradermal immunization significantly reduced the inflammatory reaction induced by influenza virus challenge in the lungs, and cGAMP-adjuvanted mRNA LNP immunization significantly improved the regional T cell immune responses.
Early immune responses induced by various adjuvanted mRNA LNPs
To investigate the adjuvanticity of cGAMP and αGC in mRNA LNPs, we determined the immune responses at the early time point and late stage after one dose of various LNPs vaccination. The T cell populations from the inguinal lymph nodes (ILNs) and spleens were analyzed on day 3 after LNPs, cGAMP-LNPs, or αGC-LNPs intradermal immunizations. Compared with naive mice, there were significantly increased CD8 and CD69 double-positive populations in ILNs after cGAMP and αGC-adjuvanted LNPs immunization (Figure 6A). No difference in CD4+CD69+ or CD8+CD69+ population was observed among the naive, LNP-, or cGAMP-LNP-immunized mouse spleens. At the same time, additional αGC significantly raised the CD69+ T cell populations in the spleens as early as day 3 (Figure 6B). Meanwhile, there were increased CD1d-tetramer and TCRβ double-positive iNKT cells in spleens from αGC-LNP-immunized mice (Figure 6C).
Figure 6.
Early immune responses induced by mRNA LNPs skin vaccination
Mice were immunized with various kinds of multivalent influenza mRNA LNPs, including LNPs, cGAMP, or αGC adjuvanted LNPs through intradermal injection. Mice were euthanized on day 3 after immunization, and the inguinal lymph nodes (ILNs), spleens, and skins were collected for analysis. (A and B) CD69-positive CD4 and CD8 populations in ILNs and spleens. (C) The iNKT populations in ILNs and spleens. (D) The DC populations in the skin after different mRNA LNPs stimulation. (E) PBMC were isolated on day 7 post-vaccination and analyzed for the CD44-positive CD4 and CD8 T cell populations. (F) The CD44-positive T cell populations in lungs at day 12 post-vaccination. The histograms were presented as mean ± SEM. Statistical significance was analyzed by F-test and two-tailed t test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; n.s. represented no significance. Data represented one experiment with n = 4 mice per group.
Skin tissue samples in the injection area were collected, and the lymphocytes were isolated to determine the changes in cell populations. Mouse dermis contains two major subsets of conventional DCs (cDCs): CD103+CD11b− (CD103+ cDCs) and CD103−CD11b+ (CD11b+ cDCs) cells.38,39 As shown in Figure 6D, there was a significantly higher percentage of CD103+ cDCs populations than CD11b+ cDCs populations (10.3% vs. 2.85%) in the dermis from naive mice. Compared with naive mice, we observed significantly increased CD11b+ cDCs in the dermis and reduced CD103+ cDCs 3 days after LNPs or cGAMP-LNPs vaccination. The lymphocytes isolated from αGC-LNP-immunized mice skins showed similar changes in cDC populations as LNP- and cGAMP-LNP-immunized mice.
We also analyzed T cell populations in PBMCs on day 7 after vaccination. CD44 is a T cell activation marker that participates in T cell differentiation and recruitment of T cells from the blood into the tissue. We found significantly increased CD44+CD4 and CD44+CD8 T cell populations in PBMC samples from LNP- or cGAMP-LNP-immunized mice. No differences were observed in αGC-LNP-immunized mice compared with naive mice (Figure 6D).
As shown in Figure 5, there were enhanced T cell immune responses in the pulmonary region after cGAMP-LNPs immunization; thus, we determined the T cell populations in the lungs at day 12 after one-dose immunization. Compared with the naive mice, LNPs and cGAMP adjuvant LNPs induced increased CD4+CD44+ and CD8+CD44+ T cell populations in the lungs (Figures 6E and 6F). Higher percentages of CD4+CD44+ T cells were detected in cGAMP or αGC adjuvanted mRNA LNP-immunized mice. These results suggested that at the early time points after cutaneous vaccination, αGC-LNPs tended to induce strong cellular immune responses in the secondary lymphoid tissue, such as spleens and lymph nodes. At the same time, cGAMP-LNPs stimulated T cell activation in ILNs and then promoted T cell activation in the blood. The significantly increased CD44+ T cell populations in the lung in the early days following mRNA LNPs immunization may play an important role in developing later cellular immune responses.
Late immune responses after mRNA LNP immunization
To study the impact of cGAMP or αGC adjuvanted LNPs formulations on immune responses at the late stages of immune responses, different groups of mice were immunized with LNPs without or with cGAMP or αGC to analyze the immune responses at week 7 after the primary immunization. Compared with the sera from naive mice, there were significantly increased Aichi-specific IgG, IgG1, and IgG2a levels in sera 7 weeks after one dose of mRNA LNPs immunization (Figure 7A). We observed lower IgG, IgG1, and IgG2a antibody titers from αGC-LNP-immunized sera. Mice were then euthanized to analyze the immune responses in the lungs and spleens. Improved IgG levels were measured in BALF from LNP- or cGAMP-LNP-immunized mice at 7 weeks after one dose of vaccine (Figure 7B).
Figure 7.
Late immune response induced by different mRNA lipid nanoparticle immunization
Sera were collected at 7 weeks after single dose of different mRNA LNPs immunizations. (A) Aichi-specific IgG, IgG1, and IgG2a endpoint titers. Mice were euthanized to collect spleens, lungs, and BALFs for analysis. (B) Aichi-specific IgG levels in BALFs. (C) Frequency of CD44+CD62L– CD4 and CD8 T cell populations in spleens were determined by flow cytometry. (D) The percentages of CD44+ CD4 and CD8 T cells in lungs. (E) and (F) The percentages of CD4+CD44+CD103+CD69+ populations in lungs. (G) Overview of immune responses after cGAMP-adjuvanted mRNA LNPs vaccination in this study. The red arrows indicated the induction of enhanced immune responses after cGAMP-LNPs immunization. The illustration was created with Biorender. The histograms were presented as mean ± SEM. Statistical significance was analyzed by F-test and two-tailed t test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. Data represented one experiment with n = 4 mice per group.
Memory T cells express high levels of CD44, which participates in the differentiation of memory T cells and regulation of memory T cell function.40,41,42 The central memory T (TCM) cells and effector memory T (TEM) cells are two kinds of memory T cell subtypes that function in secondary lymphoid tissues and peripheral tissues.43 Significantly increased CD4+CD44+CD62L– and CD8+CD44+CD62L– TEM cell populations were detected in spleens after mRNA LNPs immunization, among which the cGAMP-adjuvanted mRNA LNPs vaccination elicited higher percentages of the two populations (Figure 7C). Interestingly, cGAMP-LNPs immunization promoted significantly enhanced CD4+CD44+ and CD8+CD44+ populations in the lung, compared with no difference in inductions observed in naive, LNPs, or αGC-LNPs immunized groups (Figure 7D).
The CD44-positive T cells were further gated for the CD69 and CD103 double-positive populations to determine the lung tissue-resident memory T cells.44,45 In comparison with the naive mice and LNPs or αGC-LNPs immunized groups, the CD44+CD69+CD103+ CD4 T cells only accumulated in the lungs at the late time point after cGAMP-LNPs immunization (Figures 7E and 7F). These results suggested that the supplementation of cGAMP in mRNA LNP formulation showed more sustainable antibody responses than the addition of αGC. cGAMP-LNPs facilitated the formation of increased T cell populations in peripheral tissues and the respiratory system after vaccination, and the induction of memory T cells was supposed to be to provide rapid and efficacious protective immunity against viral infection.46,47
An overview of the immune responses induced by cGAMP-adjuvanted mRNA LNP vaccination in this study is summarized in Figure 7G. This work systemically investigated cGAMP and αGC adjuvanted mRNA LNP-elicited immune reactions and found that the cGAMP-adjuvanted mRNA LNPs promoted robust cellular immune responses in the spleen, bone marrow, and lung and enhanced the development of memory immunity in the pulmonary area. Therefore, cGAMP is a potential adjuvant for supplementing current mRNA LNP formulations.
Discussion
mRNA LNP vaccines can induce protection against various viral pathogens, such as influenza, SARS-CoV-2, and Zika virus.12,48,49 In this study, we developed a multi-component influenza mRNA LNP vaccine. The mRNAs encoded influenza HAs from two groups (H1 from Group 1 and H3 from Group2), NP, and M1. Despite the low dosage of the predominant antigens, our multivalent mRNA LNP formulation provided full protection against homologous viral infections. This study systemically investigated mRNA LNP vaccination using multivalent influenza mRNA LNPs.
The mRNA LNPs induced strong humoral and cellular immune responses after boosting immunizations. In contrast to the Th1-biased cellular immunity induced by WIVs, mRNA LNPs promoted balanced Th1/Th2 type responses, which showed high IgG2a and IgG1 levels in sera and comparably increased numbers of IFN-γ- and IL-4-secreting cells and virus-specific CD8 T cells in spleens. In addition to detecting many cytokine-secreting cells, there were significantly increased antigen-specific ASCs in spleen and bone marrow tissues after mRNA LNPs immunization. We also found that mRNA LNP intradermal immunization enhanced T cell responses in peripheral blood and lungs with significantly enhanced CD44 positive T cell populations quickly after immunization, which might indicate that mRNA LNP immunization could establish certain localized T cell memory in the pulmonary area. These mechanisms should be further investigated.
Besides fabricating multiple mRNAs into LNPs, we also explored adjuvants that could be packaged into the particles based on their characteristics, such as hydrophilicity and electricity. Hydrophilic and hydrophobic adjuvant molecules can be mixed with the mRNAs or lipids to generate mRNA LNPs with core-incorporated or surface-anchored adjuvants. cGAMP has been applied alone or in combination with other molecular adjuvants in multiple vaccine studies in recent years.18,50,51 Previous studies have detailed how cGAMP stimulates the STING-initiated downstream type I interferon signaling pathway and induces cytokine expression. The innate signaling features of cGAMP give it potential as a safe and promising adjuvant candidate.52,53
The present study encapsulated the small negatively charged hydrophilic cGAMP molecule with mRNAs into LNPs. The particulate cGAMP induced significantly higher IFN-β-promoted luciferase activity than an equivalent amount of soluble cGAMP transfection. The in vitro stimulation of BMDC consistently demonstrated the induction of type I interferon cytokines-IFN-α/β after incubating with cGAMP-adjuvanted mRNA LNPs. Similarly, enhanced αGC loading efficacy to CD1d on BMDCs was observed when αGC was encapsulated into the cationic lipid mixture.
These results indicated that LNPs could flexibly deliver mRNAs together with molecular adjuvants to synergize a comprehensive immune response. However, the cGAMP/αGC double-adjuvanted mRNA LNPs resulted in reduced cellular immune responses in spleens and the bone marrow compared with the cGAMP-adjuvanted mRNA LNPs, which was consistent with the lower IgG and IgG2a antibody levels, as well as less effective protection against homologous viral challenge. This indicated the importance of choosing an appropriate adjuvant or adjuvant combination for mRNA LNP vaccines.
Notably, cGAMP is likely to improve vaccine safety by limiting systemic proinflammation.50 Supplementation with cGAMP further increased the CD4 and CD8 T cell populations in BALF and the NP147-155-tetramer + CD8 T cells in the lungs after viral infection. Compared with cGAMP, αGC-adjuvanted LNPs stimulated broader cellular responses as early as 3 days after immunization with increased active T cells in the spleens and ILNs and expanded iNKT cell populations in the spleens. Activated iNKT cells rapidly secrete Th1 and Th2 cytokines to facilitate DC maturation, germinal center B cell responses,54 and cytotoxic CD8+ T cell activation. However, iNKT cells can also exhibit potent cytotoxicity, which could impair effective immune responses and diminish the protection against viral infection.55,56
Many studies have demonstrated that enhanced viral-specific cellular immune responses are essential for protection against viral infection, in addition to facilitating viral clearance and regulation of antibody response, robust T cell responses influence the development of memory immunity.57,58,59 The pleiotropic functions of CD4 T cells in recruitment, activation, and maintenance of innate immune cells, B cells, and CD8 T cells in respiratory tissues have been further illustrated.60,61 According to our studies, cGAMP-adjuvanted mRNA LNPs immunization induced significantly stronger cellular immune responses.
Different types of vaccine strategies and vectors were studied to generate pulmonary TRM through respiratory system deliveries, like intranasal administration to protect against respiratory viral infections by evoking rapid responses at mucosal sites, promoting viral clearance, and reducing disease severity.31,62,63 A key difference from previous works in this study provides evidence for the accumulation of CD4 T cells in lungs after cGAMP-adjuvanted mRNA LNP skin vaccination. The higher percentage of CD4+CD44+CD103+CD69+ T cell population induced by cGAMP-adjuvanted mRNA LNP cutaneous immunization indicated that the combination of cGAMP LNP formulation and intradermal immunization route in this study might promote the activation and circulation of T cells to nonlymphoid tissue, such as the lung. Previous studies have found that cutaneous immunization could promote mucosal immune responses.64,65 Whether the enhanced T cell responses confer durable immune protection and if an increased memory T cell population evolves in the setting of the present study are subjects of future exploration.
Therefore, we have found that cGAMP is an effective adjuvant for mRNA LNPs when applied in intradermal vaccination. These dose-sparing cGAMP-adjuvanted mRNA vaccines could be applied to other more relevant and convenient vaccination strategies, such as biodegradable microneedle patches (MNPs).66 The MNP skin vaccination attenuates the tissue damage-related inflammation caused by intradermal or intramuscular injection. Overall, this study illustrated that cGAMP-adjuvanted mRNA LNPs are a promising vaccine candidate for future vaccine development.
Materials and methods
Ethics statement
This study was performed following the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The animal facility at Georgia State University is fully accredited by the American Association for Assessment and Accreditation of Laboratory Animal Care. All the animal studies were approved by the Georgia State University Institutional Animal Care and Use Committee under protocol No. A19025.
Viruses, cells, and reagents
Mouse-adapted influenza A/PR/8/34 (H1N1, PR8), A/Aichi/2/68 (H3N2, Aichi), and A/Philippines/2/1982 (H3N2, Phi) were expanded from infected mice, and LD50 values were determined. PR8 and Aichi, used to produce whole inactivated viruses, were amplified in embryonated chicken eggs. The reassortant A/Shanghai/2013 H7N9 (rSH) influenza virus with the PR8 backbone was amplified in eggs and was used in the mice challenge studies.67 Madin-Darby canine kidney (MDCK, ATCC CCL-34) cells and 293T cells were cultured in DMEM with 10% FBS. Sf9 cells (ATCC, CRL-1711) were cultured in ESF 921 insect cell culture medium.
As previously described, BMDCs were isolated and cultured.68 Briefly, 10 × 106 bone marrow cells were cultured in a 10-cm dish in 10 mL of completed medium (cRPMI, RPMI 1640 supplemented with glutamine, penicillin, streptomycin, 2-mercaptoethanol, HEPES, non-essential amino acid, sodium pyruvate, and 10% FBS) with GM-CSF (20 ng mL−1). Cells were cultured at 37°C for 2 days. Half of the medium was replaced with a new medium with GM-CSF (20 ng mL−1) on day 2. On day 3, the entire medium was replaced with a fresh complete medium supplemented with GM-CSF (20 ng mL−1) and then cultured for another 3 days. On day 6, non-adherent and loosely adherent cells were harvested by centrifuging at 250x g for 8 min.
DOTMA, DOPE, and DMG-PEG 2000 were purchased from Avanti Polar Lipids. Cholesterol and 2′3′-cGAMP were bought from Sigma and InvivoGen, respectively. PE-conjugated H-2K(d) Influenza An NP 147-155 TYQRTRALV tetramer (IEDB ID 67436) and mouse CD1d PBS-57 were obtained from NIH Tetramer Core Facility at Emory University. The antibodies used in flow cytometry analysis were purchased from BD Biosciences and Biolegend. The recombinant hemagglutinin proteins H4 (NR-51128, A/mallard/Alberta/455/2015 [H4N6]), H3 (NR-19241, A/New York/55/2004 [H3N2]), and H1 (NR-51668, A/California/07/2009 [H1N1] pdm09) were obtained from BEI Resources. As previously described, the recombinant Aichi H3, PR8 H1, and PR8 NP proteins were purified from Sf9 cells using the Bac-to-Bac baculovirus expression system (Invitrogen).67 The influenza A H1N1 (A/Brevig Mission/1/1918) matrix protein (M1) was purchased from Sinobiology.
RNA production
Encoding genes of influenza HAs from A/PR/8/34 (H1N1, PR8), A/Aichi/2/68 (H3N2, Aichi), NP, and M1 from A/PR/8/34 (H1N1, PR8) were cloned into modified pMRNAxp mRNA Express Vector (SBI system biosciences). The mRNA production plasmids were linearized by PciI single enzyme digestion. The DNA templates were purified using phenol:ChCl3:IAA 25:24:1 (Cat# AM9730, Ambion) and used for mRNA in vitro transcription. mRNAs were produced using T7 mRNA polymerase (MEGAscript T7 kit, Invitrogen). During the in vitro transcription, N1-methylpseudouridine-5′-triphosphate (APExBIO) was used to generate modified nucleoside-containing mRNAs instead of UTP (uridine 5′-triphosphate). The transcribed mRNAs containing 120-nt poly(A) tails were furtherly capped using ScriptCap Cap 1 Capping System (CELLSCRIPT, Cat. No. C-SCCS1710). mRNAs were purified using a MEGAclear transcription clean-up kit from Invitrogen. All the mRNAs were analyzed by denaturing or native agarose gel electrophoresis and were stored at −80°C. The mRNAs were transfected into 293T cells with Lipofectamine MessengerMAX (Thermo Fisher Scientific), and protein samples were collected 12 h and 24 h post-transfection to determine the mRNA expression by western blot.
Fabrication and characterization of mRNA LNPs
Lipids were dissolved in ethanol at molar ratios of 50:10:38.5:1.5 (DOTMA:DOPE:cholesterol:DMG-PEG 2000). The lipid mixture was mixed with mRNAs diluting in 25 mM sodium citrate (pH 4.0) at a volume ratio of 3:1 (aqueous:ethanol) using a microfluidic mixer (Precision Nanosystems). The mRNA to total lipid ratio was 0.05 (wt/wt) for each formulation. For multivalent mRNA lipid nanoparticles, 20 μg of total mRNAs containing 5 μg of each Aichi-HA3, PR8-HA1, PR8-NP, and PR8-M1 mRNAs were fabricated together into lipid nanoparticles (LNPs). 2 μg of cGAMP was mixed with 20 μg of mRNAs, and 2.5% of αGC was combined with lipid fractions to obtain cGAMP or αGC-adjuvanted multivalent mRNA LNPs. The sizes and zeta potentials of mRNA LNPs were measured by dynamic light scattering analysis with Malvern Zetasizer. The morphology of LNPs was characterized by transmission electron microscopy.
The mRNA LNPs were treated with 0.8% Triton X-100 and vortexed for 5 min to disrupt the lipid structure. The total and unencapsulated mRNA concentrations were measured by RiboGreen RNA reagent (Invitrogen). The encapsulation efficiency (EE%) was calculated as (total mRNA – unencapsulated mRNA)/total mRNA × 100%. The mRNA LNPs were transfected into 293T, and then we measured the protein expression at 12 h and 24 h by western blot. Cultured BMDCs were stimulated with mRNA LNPs for 24 h; the supernatants were collected to analyze the expression of cytokines. As previously described, the levels of IFN-α, IFN-β, and IL-12 were measured by sandwich cytokine ELISA.66 Briefly, the Nunc Maxisorp ELISA plates were coated with 4 μg mL−1 of captured antibodies at 4°C overnight. Fifty microliters of BMDC cultured supernatant were added into coated wells and incubated at 37°C for 2 hours followed by the incubation of biotin-conjugated detection antibodies and HRP-conjugated streptavidin. Serial dilutions of the cytokine standards were included to generate standard curves.
293T cells were transfected with STING (Plasmid #124262) and IFN-β-luc (Plasmid #102597) or IFN-γ-luc (Plasmid #17599) promoter-reporter plasmids that were purchased from Addgene. 24 h post-transfection, cells were split into 24-well plates and then transfected with soluble cGAMP or particulate cGAMP in lipids. The luciferase activities were measured 6 h post-transfection using the Bright-Glo reagent (Promega). To measure the ability to form αGC:CD1d complex, soluble αGC and αGC– particles were incubated with BMDCs for 24 h. The BMDCs were collected 24 h later and stained with PE-conjugated anti-mouse αGC:CD1d complex antibody and CD11c-Percp/Cy5.5 (Biolegend) to evaluate the percentage of αGC:CD1d complex positive DCs by flow cytometry. The lipid formulation for fabrication of mRNA LNPs in this study was used to synthesize particulate cGAMP and αGC.
Immunization and challenges in mice
For different kinds of multivalent influenza mRNA LNP vaccination, 6- to 8-week-old female BALB/c mice (Envigo) were intradermally injected with LNPs containing Aichi-HA3 (5 μg), PR8-HA1 (5 μg), PR8-NP (5 μg), and PR8-M1 (5 μg) or mRNA LNPs incorporated with 2 μg of cGAMP or cGAMP (2 μg)/αGC (2.5%). One group of naive mice was used as a control. Mice received priming and boosting immunizations at a 4-week interval. At week 4 after boosting immunization, mice were challenged with 5x LD50 PR8, 3x LD50 Phi, 30x LD50 Aichi, or 3x LD50 rSH. Mouse body weights were monitored daily for 14 days. For Aichi-LNPs immunization, 10 μg of Aichi-HA3 mRNA was fabricated into LNPs. For the WIV immunized group, mice were immunized with WIVs containing 1 μg of A/Aichi and 1 μg of A/PR8.
Humoral immune responses
Blood samples were collected 3 weeks and 7 weeks after priming immunization and 3 weeks after boosting immunization. The levels of Aichi, PR8, NP, and M1 specific IgG and IgG isotype (IgG1 and IgG2a) antibody levels were measured by ELISA using 4 μg mL−1 of purified whole inactivated Aichi and PR8 viruses or purified NP or M1 proteins as coating antigens, respectively. The recombinant hemagglutinin proteins H4 (NR-51128, A/mallard/Alberta/455/2015 [H4N6]), H3 (NR-19241, A/New York/55/2004 [H3N2]), and H1 (NR-51668, A/California/07/2009 [H1N1] pdm09) were also used as coating antigens to determine the antigen-specific IgG levels by ELISA. Mice BALFs were collected 6 days after challenge and 7 weeks after primary immunization, and the levels of virus-specific IgG were measured by ELISA.
Cellular immune responses
Mice were sacrificed at week 4 post-immunization. As previously described, the spleens and the bone marrow were collected and processed into single-cell suspensions with cRPMI media for the ELISPOT assays.69 To determine antigen-specific IL-4 and IFN-γ-secreting cells, 1 × 106 splenocytes were seeded with A/Aichi, A/PR8 whole inactivated viruses, or NP peptides (BEI Resources, NR-2611) as stimulators into 96-well filtration plates (Millipore), which were pre-coated with 4 μg mL−1 of anti-mouse IL-4 or IFN-γ capture antibodies (Biolegend). The plates were incubated at 37°C for 48 h and then incubated with biotin-conjugated IL-4 or IFN-γ detection antibodies and HRP-streptavidin (BioLegend), followed by KPL TrueBlue substrate staining. Results were measured by Bioreader-6000-E (BIOSYSTEM). To analyze virus-specific ASCs in spleens and bone marrow, 1 × 106 cells were seeded into 96-well filtration plates pre-coated with 4 μg mL−1 of A/Aichi, A/PR8 WIVs. 18 h after incubation, plates were washed and incubated with HRP-conjugated goat anti-mouse IgG. The wells were stained, and spots were counted by Bioreader-6000-E.
BALFs and lung tissues were also collected 6 days after virus infection. The cells from BALFs were collected by centrifugation at 500x g for 10 min and stained with CD4-Percp, CD8-FITC, and CD16/32 antibodies in staining buffer (PBS + 2%FBS) for T cell population analysis by flow cytometry. Lung tissues were processed with cRPMI media through a 70-μm cell strainer and then centrifuged at 500x g for 10 min. The pellets were resuspended in 4 mL PBS and loaded gently on 4 mL Histopaque-1803 in 15-mL conical tubes. The tubes were centrifuged at 400x g for 30 min with the lowest speed-up and brake-off. The lymphocyte fraction in the middle was carefully harvested and washed with fresh PBS. The pellets were resuspended in the staining buffer and stained with PE-NP147-155-tetramer, CD8-FITC, CD4-Percp, CD44-PE, and CD16/32 antibodies to determine the T cell populations by flow cytometry. The tissue-resident memory T cell populations were determined by staining with CD45-PE, CD4-Percp, CD8-APC, CD44-BV421, CD103-FITC, CD69-PE/Cy7, and CD16/32.
A similar method was applied to isolate PBMCs from mouse blood using Histopaque-1803. Briefly, the mouse blood was diluted with PBS and gently layered on the same volume of Histopaque-1083. The tubes were centrifuged at 400x g for 30 min at room temperature with brake-off. The isolated PBMCs were stained with CD4-Percp, CD8-FITC, CD44-PE, and CD16/32 antibodies to determine the T cell populations in the blood. The isolation of mouse skin lymphocytes was performed following a previously described protocol.70 The cells were stained with MHCII-PE, CD11c-APC, CD11b-Percp/Cy5.5, CD103-FITC, and CD16/32 to analyze DC populations in the skin by flow cytometry. One million cells from ILNs or spleens were centrifuged, resuspended in staining buffer, and then stained with CD4-Percp, CD8-FITC, CD69-PE, and CD16/32 for T cell analysis. Splenocytes were stained with CD45-PE, CD4-Percp, CD8-APC, CD44-BV421, CD62L-PE, and CD16/32 to determine the T cell populations by flow cytometry.
Histological analysis and lung virus titers
The murine lung tissues were collected 6 days after viral infection. Parts of lungs from different immunization groups were separated for histological analysis as previously described.71 Briefly, tissues were washed with PBS and then fixed with 10% neutral buffered formalin at 4°C overnight. These tissues were embedded in paraffin after dehydration. The paraffin sections were stained with hematoxylin and eosin. The rest of the lung samples were homogenized with PBS, and the supernatants containing the viruses were collected by centrifugation at 500x g for 10 min. After that, the lung supernatants were 10-fold serially diluted, added into MDCK cells (1 × 105/well) in 96-well plates, and then cultured for 5 days. After incubation, the HA titers in the MDCKs supernatants from each well were determined, and the lung viral titers were calculated by the Reed-München method.
Statistical analysis
The data were represented by means with the standard error of the mean (SEM). The p values were assigned by calculating the significant differences between two groups by F-test and a two-tailed Student’s t test. p values less than 0.05 were considered to be statistically significant.
Acknowledgments
This work was supported by the US NIH/National Institute of Allergy and Infectious Diseases under Grants R01AI101047, R01AI116835, and R01AI143844 to B.-Z.W. The electron microscopy study was performed in part at the Georgia Institute of Technology for Electronics and Nanotechnology, a member of the National Nanotechnology Coordinated Infrastructure supported by the NSF (Grant ECCS-1542174). The content in this study is solely our responsibility and does not necessarily represent the official views of the funders. The graphical abstract was created with BioRender.
Author contributions
W.Z, L.W., C.D., Y.W., J.K., and Y.M. conducted the experiments; B.-Z.W. and W.Z. designed the experiments; W.Z. wrote the original draft, and B.-Z. W., W.Z., and G.X.G. reviewed and edited the manuscript.
Declaration of interests
The authors have no financial or commercial conflict of interest.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2022.10.024
Supplemental information
Document S2. Article plus supplemental information
Data availability
The data that support the findings in this study are available on Mendeley Data: https://doi.org/10.17632/jm49hvbh8k.2.
References
- 1.Allen J.D., Ross T.M. H3N2 influenza viruses in humans: viral mechanisms, evolution, and evaluation. Hum. Vaccines Immunother. 2018;14:1840–1847. doi: 10.1080/21645515.2018.1462639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petrova V.N., Russell C.A. The evolution of seasonal influenza viruses. Nat. Rev. Microbiol. 2018;16:60. doi: 10.1038/nrmicro.2017.146. [DOI] [PubMed] [Google Scholar]
- 3.Zhang C., Maruggi G., Shan H., Li J. Advances in mRNA vaccines for infectious diseases. Front. Immunol. 2019;10:594. doi: 10.3389/fimmu.2019.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines - a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gomez-Aguado I., Rodriguez-Castejon J., Vicente-Pascual M., Rodriguez-Gascon A., Solinis M.A., Del Pozo-Rodriguez A. Nanomedicines to deliver mRNA: state of the art and future perspectives. Nanomaterials. 2020;10 doi: 10.3390/nano10020364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alameh M.G., Tombacz I., Bettini E., Lederer K., Sittplangkoon C., Wilmore J.R., Gaudette B.T., Soliman O.Y., Pine M., Hicks P., et al. Lipid nanoparticles enhance the efficacy of mRNA and protein subunit vaccines by inducing robust T follicular helper cell and humoral responses. Immunity. 2021;54:2877–2892.e7. doi: 10.1016/j.immuni.2021.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole S.L., Cybulski V., Whitacre M., Lathrop S.K., Bazin-Lee H., Burkhart D., Evans J.T. Influenza vaccines using liposomal formulations of toll-like receptor (TLR) 7/8 and 4 agonists as adjuvants. J. Immunol. 2020;204 [Google Scholar]
- 8.Shirai S., Shibuya M., Kawai A., Tamiya S., Munakata L., Omata D., Suzuki R., Aoshi T., Yoshioka Y. Lipid nanoparticles potentiate CpG-oligodeoxynucleotide-based vaccine for influenza virus. Front. Immunol. 2019;10:3018. doi: 10.3389/fimmu.2019.03018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Perez Marc G., Moreira E.D., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pardi N., Parkhouse K., Kirkpatrick E., McMahon M., Zost S.J., Mui B.L., Tam Y.K., Kariko K., Barbosa C.J., Madden T.D., et al. Nucleoside-modified mRNA immunization elicits influenza virus hemagglutinin stalk-specific antibodies. Nat. Commun. 2018;9:3361. doi: 10.1038/s41467-018-05482-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freyn A.W., Ramos da Silva J., Rosado V.C., Bliss C.M., Pine M., Mui B.L., Tam Y.K., Madden T.D., de Souza Ferreira L.C., Weissman D., et al. A multi-targeting, nucleoside-modified mRNA influenza virus vaccine provides broad protection in mice. Mol. Ther. 2020;28:1569–1584. doi: 10.1016/j.ymthe.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pardi N., Hogan M.J., Naradikian M.S., Parkhouse K., Cain D.W., Jones L., Moody M.A., Verkerke H.P., Myles A., Willis E., et al. Nucleoside-modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. J. Exp. Med. 2018;215:1571–1588. doi: 10.1084/jem.20171450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhuang X., Qi Y., Wang M., Yu N., Nan F., Zhang H., Tian M., Li C., Lu H., Jin N. mRNA Vaccines Encoding the HA Protein of Influenza A H1N1 Virus Delivered by Cationic Lipid Nanoparticles Induce Protective Immune Responses in Mice. Vaccines. 2020;8 doi: 10.3390/vaccines8010123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Islam M.A., Rice J., Reesor E., Zope H., Tao W., Lim M., Ding J., Chen Y., Aduluso D., Zetter B.R., et al. Adjuvant-pulsed mRNA vaccine nanoparticle for immunoprophylactic and therapeutic tumor suppression in mice. Biomaterials. 2021;266:120431. doi: 10.1016/j.biomaterials.2020.120431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee K., Kim S.Y., Seo Y., Kim M.H., Chang J., Lee H. Adjuvant incorporated lipid nanoparticles for enhanced mRNA-mediated cancer immunotherapy. Biomater. Sci. 2020;8:1101–1105. doi: 10.1039/c9bm01564g. [DOI] [PubMed] [Google Scholar]
- 17.Wang J., Li P., Wu M.X. Natural STING agonist as an “ideal” adjuvant for cutaneous vaccination. J. Invest. Dermatol. 2016;136:2183–2191. doi: 10.1016/j.jid.2016.05.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vassilieva E.V., Li S., Korniychuk H., Taylor D.M., Wang S., Prausnitz M.R., Compans R.W. cGAMP/saponin adjuvant combination improves protective response to influenza vaccination by microneedle patch in an aged mouse model. Front. Immunol. 2020;11:583251. doi: 10.3389/fimmu.2020.583251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ko S.Y., Ko H.J., Chang W.S., Park S.H., Kweon M.N., Kang C.Y. alpha-galactosylceramide can act as a nasal vaccine adjuvant inducing protective immune responses against viral infection and tumor. J. Immunol. 2005;175:3309–3317. doi: 10.4049/jimmunol.175.5.3309. [DOI] [PubMed] [Google Scholar]
- 20.Lindqvist M., Persson J., Thorn K., Harandi A.M. The mucosal adjuvant effect of alpha-galactosylceramide for induction of protective immunity to sexually transmitted viral infection. J. Immunol. 2009;182:6435–6443. doi: 10.4049/jimmunol.0900136. [DOI] [PubMed] [Google Scholar]
- 21.Qin Y., Oh S., Lim S., Shin J.H., Yoon M.S., Park S.H. Invariant NKT cells facilitate cytotoxic T-cell activation via direct recognition of CD1d on T cells. Exp. Mol. Med. 2019;51 doi: 10.1038/S12276-019-0329-9. Artn 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghinnagow R., Cruz L.J., Macho-Fernandez E., Faveeuw C., Trottein F. Enhancement of adjuvant functions of natural killer T cells using nanovector delivery systems: application anticancer immune therapy. Front. Immunol. 2017;8 doi: 10.3389/Fimmu.2017.00879. Artn 879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dolen Y., Kreutz M., Gileadi U., Tel J., Vasaturo A., van Dinther E.A., van Hout-Kuijer M.A., Cerundolo V., Figdor C.G. Co-delivery of PLGA encapsulated invariant NKT cell agonist with antigenic protein induce strong T cell-mediated antitumor immune responses. OncoImmunology. 2016;5:e1068493. doi: 10.1080/2162402X.2015.1068493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thapa P., Zhang G.D., Xia C.F., Gelbard A., Overwijk W.W., Liu C.W., Hwu P., Chang D.Z., Courtney A., Sastry J.K., et al. Nanoparticle formulated alpha-galactosylceramide activates NKT cells without inducing anergy. Vaccine. 2009;27:3484–3488. doi: 10.1016/j.vaccine.2009.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sainz V., Moura L.I.F., Peres C., Matos A.I., Viana A.S., Wagner A.M., Vela Ramirez J.E., T S.B., Gaspar M., Brocchini S., et al. alpha-Galactosylceramide and peptide-based nano-vaccine synergistically induced a strong tumor suppressive effect in melanoma. Acta Biomater. 2018;76:193–207. doi: 10.1016/j.actbio.2018.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Z.T., Zarnitsyna V.I., Lowen A.C., Weissman D., Koelle K., Kohlmeier J.E., Antia R. Why are CD8 T cell epitopes of human influenza A virus conserved? J. Virol. 2019;93 doi: 10.1128/JVI.01534-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Puksuriwong S., Ahmed M.S., Sharma R., Krishnan M., Leong S., Lambe T., McNamara P.S., Gilbert S.C., Zhang Q. Modified vaccinia ankara-vectored vaccine expressing nucleoprotein and matrix protein 1 (M1) activates mucosal M1-specific T-cell immunity and tissue-resident memory T cells in human nasopharynx-associated lymphoid tissue. J. Infect. Dis. 2020;222:807–819. doi: 10.1093/infdis/jiz593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen G.B., Yang X.B., Ko A., Ga M.M., Zhang Y.Q., Shi A., Sun X.P., Mariuzza R.A., Weng N.P. Distinct features of human CD8 T cell TCR repertoire specific to influenza A virus matrix protein M1. J. Immunol. 2016;196 [Google Scholar]
- 29.McMahon M., Asthagiri Arunkumar G., Liu W.C., Stadlbauer D., Albrecht R.A., Pavot V., Aramouni M., Lambe T., Gilbert S.C., Krammer F. Vaccination with viral vectors expressing chimeric hemagglutinin, NP and M1 antigens protects ferrets against influenza virus challenge. Front. Immunol. 2019;10:2005. doi: 10.3389/fimmu.2019.02005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asthagiri Arunkumar G., McMahon M., Pavot V., Aramouni M., Ioannou A., Lambe T., Gilbert S., Krammer F. Vaccination with viral vectors expressing NP, M1 and chimeric hemagglutinin induces broad protection against influenza virus challenge in mice. Vaccine. 2019;37:5567–5577. doi: 10.1016/j.vaccine.2019.07.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zens K.D., Chen J.K., Farber D.L. Vaccine-generated lung tissue-resident memory T cells provide heterosubtypic protection to influenza infection. JCI Insight. 2016;1 doi: 10.1172/jci.insight.85832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koutsakos M., Illing P.T., Nguyen T.H.O., Mifsud N.A., Crawford J.C., Rizzetto S., Eltahla A.A., Clemens E.B., Sant S., Chua B.Y., et al. Human CD8(+) T cell cross-reactivity across influenza A, B and C viruses. Nat. Immunol. 2019;20:613–625. doi: 10.1038/s41590-019-0320-6. [DOI] [PubMed] [Google Scholar]
- 33.Macho-Fernandez E., Cruz L.J., Ghinnagow R., Fontaine J., Bialecki E., Frisch B., Trottein F., Faveeuw C. Targeted delivery of alpha-galactosylceramide to CD8alpha+ dendritic cells optimizes type I NKT cell-based antitumor responses. J. Immunol. 2014;193:961–969. doi: 10.4049/jimmunol.1303029. [DOI] [PubMed] [Google Scholar]
- 34.Jensen S.S., Gad M. Differential induction of inflammatory cytokines by dendritic cells treated with novel TLR-agonist and cytokine based cocktails: targeting dendritic cells in autoimmunity. J. Inflamm. 2010;7:37. doi: 10.1186/1476-9255-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McNab F., Mayer-Barber K., Sher A., Wack A., O'Garra A. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015;15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Macatonia S.E., Hosken N.A., Litton M., Vieira P., Hsieh C.S., Culpepper J.A., Wysocka M., Trinchieri G., Murphy K.M., O'Garra A. Dendritic cells produce IL-12 and direct the development of Th1 cells from naive CD4+ T cells. J. Immunol. 1995;154:5071–5079. [PubMed] [Google Scholar]
- 37.Heufler C., Koch F., Stanzl U., Topar G., Wysocka M., Trinchieri G., Enk A., Steinman R.M., Romani N., Schuler G. Interleukin-12 is produced by dendritic cells and mediates T helper 1 development as well as interferon-gamma production by T helper 1 cells. Eur. J. Immunol. 1996;26:659–668. doi: 10.1002/eji.1830260323. [DOI] [PubMed] [Google Scholar]
- 38.Merad M., Sathe P., Helft J., Miller J., Mortha A. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu. Rev. Immunol. 2013;31:563–604. doi: 10.1146/annurev-immunol-020711-074950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sichien D., Lambrecht B.N., Guilliams M., Scott C.L. Development of conventional dendritic cells: from common bone marrow progenitors to multiple subsets in peripheral tissues. Mucosal Immunol. 2017;10:831–844. doi: 10.1038/mi.2017.8. [DOI] [PubMed] [Google Scholar]
- 40.Eberl G., Brawand P., MacDonald H.R. Selective bystander proliferation of memory CD4+ and CD8+ T cells upon NK T or T cell activation. J. Immunol. 2000;165:4305–4311. doi: 10.4049/jimmunol.165.8.4305. [DOI] [PubMed] [Google Scholar]
- 41.Baaten B.J., Li C.R., Bradley L.M. Multifaceted regulation of T cells by CD44. Commun. Integr. Biol. 2010;3:508–512. doi: 10.4161/cib.3.6.13495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baaten B.J., Tinoco R., Chen A.T., Bradley L.M. Regulation of antigen-experienced T cells: lessons from the quintessential memory marker CD44. Front. Immunol. 2012;3:23. doi: 10.3389/fimmu.2012.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilk M.M., Misiak A., McManus R.M., Allen A.C., Lynch M.A., Mills K.H.G. Lung CD4 tissue-resident memory T cells mediate adaptive immunity induced by previous infection of mice with bordetella pertussis. J. Immunol. 2017;199:233–243. doi: 10.4049/jimmunol.1602051. [DOI] [PubMed] [Google Scholar]
- 44.Zheng M.Z.M., Wakim L.M. Tissue resident memory T cells in the respiratory tract. Mucosal Immunol. 2022;15:379–388. doi: 10.1038/s41385-021-00461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu J., Madi A., Mieg A., Hotz-Wagenblatt A., Weisshaar N., Ma S., Mohr K., Schlimbach T., Hering M., Borgers H., Cui G. T cell factor 1 suppresses CD103+ lung tissue-resident memory T cell development. Cell Rep. 2020;31:107484. doi: 10.1016/j.celrep.2020.03.048. [DOI] [PubMed] [Google Scholar]
- 46.Teijaro J.R., Turner D., Pham Q., Wherry E.J., Lefrancois L., Farber D.L. Cutting edge: tissue-retentive lung memory CD4 T cells mediate optimal protection to respiratory virus infection. J. Immunol. 2011;187:5510–5514. doi: 10.4049/jimmunol.1102243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pizzolla A., Wakim L.M. Memory T cell dynamics in the lung during influenza virus infection. J. Immunol. 2019;202:374–381. doi: 10.4049/jimmunol.1800979. [DOI] [PubMed] [Google Scholar]
- 48.Richner J.M., Himansu S., Dowd K.A., Butler S.L., Salazar V., Fox J.M., Julander J.G., Tang W.W., Shresta S., Pierson T.C., et al. Modified mRNA vaccines protect against Zika virus infection. Cell. 2017;169:176. doi: 10.1016/j.cell.2017.03.016. [DOI] [PubMed] [Google Scholar]
- 49.Pilkington E.H., Suys E.J.A., Trevaskis N.L., Wheatley A.K., Zukancic D., Algarni A., Al-Wassiti H., Davis T.P., Pouton C.W., Kent S.J., Truong N.P. From influenza to COVID-19: lipid nanoparticle mRNA vaccines at the frontiers of infectious diseases. Acta Biomater. 2021;131:16–40. doi: 10.1016/j.actbio.2021.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chauveau L., Bridgeman A., Tan T.K., Beveridge R., Frost J.N., Rijal P., Pedroza-Pacheco I., Partridge T., Gilbert-Jaramillo J., Knight M.L., et al. Inclusion of cGAMP within virus-like particle vaccines enhances their immunogenicity. EMBO Rep. 2021;22:e52447. doi: 10.15252/embr.202152447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J., Li P., Yu Y., Fu Y., Jiang H., Lu M., Sun Z., Jiang S., Lu L., Wu M.X. Pulmonary surfactant-biomimetic nanoparticles potentiate heterosubtypic influenza immunity. Science. 2020;367 doi: 10.1126/science.aau0810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cai X., Chiu Y.H., Chen Z.J. The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Molecular cell. 2014;54:289–296. doi: 10.1016/j.molcel.2014.03.040. [DOI] [PubMed] [Google Scholar]
- 53.Ritchie C., Li L. cGAMP as an adjuvant in antiviral vaccines and cancer immunotherapy. Biochemistry. 2020;59:1713–1715. doi: 10.1021/acs.biochem.0c00226. [DOI] [PubMed] [Google Scholar]
- 54.Gaya M., Barral P., Burbage M., Aggarwal S., Montaner B., Navia A.W., Aid M., Tsui C., Maldonado P., Nair U., et al. Initiation of antiviral B cell immunity relies on innate signals from spatially positioned NKT cells. Cell. 2018;172:517. doi: 10.1016/j.cell.2017.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bassiri H., Das R., Guan P., Barrett D.M., Brennan P.J., Banerjee P.P., Wiener S.J., Orange J.S., Brenner M.B., Grupp S.A., Nichols K.E. iNKT cell cytotoxic responses control T-lymphoma growth in vitro and in vivo. Cancer Immunol. Res. 2014;2:59–69. doi: 10.1158/2326-6066.CIR-13-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wingender G., Krebs P., Beutler B., Kronenberg M. Antigen-specific cytotoxicity by invariant NKT cells in vivo is CD95/CD178-dependent and is correlated with antigenic potency. J. Immunol. 2010;185:2721–2729. doi: 10.4049/jimmunol.1001018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Panagioti E., Klenerman P., Lee L.N., van der Burg S.H., Arens R. Features of effective T cell-inducing vaccines against chronic viral infections. Front. Immunol. 2018;9:276. doi: 10.3389/fimmu.2018.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Z., Yang X., Zhong J., Zhou Y., Tang Z., Zhou H., He J., Mei X., Tang Y., Lin B., et al. Exposure to SARS-CoV-2 generates T-cell memory in the absence of a detectable viral infection. Nat. Commun. 2021;12:1724. doi: 10.1038/s41467-021-22036-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moss P. The T cell immune response against SARS-CoV-2. Nat. Immunol. 2022;23:186–193. doi: 10.1038/s41590-021-01122-w. [DOI] [PubMed] [Google Scholar]
- 60.Swain S.L., McKinstry K.K., Strutt T.M. Expanding roles for CD4(+) T cells in immunity to viruses. Nat. Rev. Immunol. 2012;12:136–148. doi: 10.1038/nri3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pruner K.B., Pepper M. Local memory CD4 T cell niches in respiratory viral infection. J. Exp. Med. 2021;218 doi: 10.1084/jem.20201733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morabito K.M., Ruckwardt T.R., Redwood A.J., Moin S.M., Price D.A., Graham B.S. Intranasal administration of RSV antigen-expressing MCMV elicits robust tissue-resident effector and effector memory CD8+ T cells in the lung. Mucosal Immunol. 2017;10:545–554. doi: 10.1038/mi.2016.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hassan A.O., Kafai N.M., Dmitriev I.P., Fox J.M., Smith B.K., Harvey I.B., Chen R.E., Winkler E.S., Wessel A.W., Case J.B., et al. A single-dose intranasal Chad vaccine protects upper and lower respiratory tracts against SARS-CoV-2. Cell. 2020;183:169–184.e13. doi: 10.1016/j.cell.2020.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lawson L.B., Clements J.D., Freytag L.C. Mucosal immune responses induced by transcutaneous vaccines. Curr. Top. Microbiol. Immunol. 2012;354:19–37. doi: 10.1007/82_2010_113. [DOI] [PubMed] [Google Scholar]
- 65.Belyakov I.M., Hammond S.A., Ahlers J.D., Glenn G.M., Berzofsky J.A. Transcutaneous immunization induces mucosal CTLs and protective immunity by migration of primed skin dendritic cells. The Journal of clinical investigation. 2004;113:998–1007. doi: 10.1172/JCI20261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu W., Li S., Wang C., Yu G., Prausnitz M.R., Wang B.Z. Enhanced immune responses conferring cross-protection by skin vaccination with a tri-component influenza vaccine using a microneedle patch. Front. Immunol. 2018;9:1705. doi: 10.3389/fimmu.2018.01705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Deng L., Mohan T., Chang T.Z., Gonzalez G.X., Wang Y., Kwon Y.M., Kang S.M., Compans R.W., Champion J.A., Wang B.Z. Double-layered protein nanoparticles induce broad protection against divergent influenza A viruses. Nat. Commun. 2018;9:359. doi: 10.1038/s41467-017-02725-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Helft J., Bottcher J., Chakravarty P., Zelenay S., Huotari J., Schraml B.U., Goubau D., Reis e Sousa C. GM-CSF mouse bone marrow cultures comprise a heterogeneous population of CD11c(+)MHCII(+) macrophages and dendritic cells. Immunity. 2015;42:1197–1211. doi: 10.1016/j.immuni.2015.05.018. [DOI] [PubMed] [Google Scholar]
- 69.Zhu W., Pewin W., Wang C., Luo Y., Gonzalez G.X., Mohan T., Prausnitz M.R., Wang B.Z. A boosting skin vaccination with dissolving microneedle patch encapsulating M2e vaccine broadens the protective efficacy of conventional influenza vaccines. J. Contr. Release. 2017;261:1–9. doi: 10.1016/j.jconrel.2017.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Benck C.J., Martinov T., Fife B.T., Chatterjea D. Isolation of infiltrating leukocytes from mouse skin using enzymatic digest and gradient separation. J. Vis. Exp. 2016:e53638. doi: 10.3791/53638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dong C., Wang Y., Gonzalez G.X., Ma Y., Song Y., Wang S., Kang S.M., Compans R.W., Wang B.Z. Intranasal vaccination with influenza HA/GO-PEI nanoparticles provides immune protection against homo- and heterologous strains. Prod. of the Natl. Acad. of Sci. of the USA. 2021;118 doi: 10.1073/pnas.2024998118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Document S2. Article plus supplemental information
Data Availability Statement
The data that support the findings in this study are available on Mendeley Data: https://doi.org/10.17632/jm49hvbh8k.2.