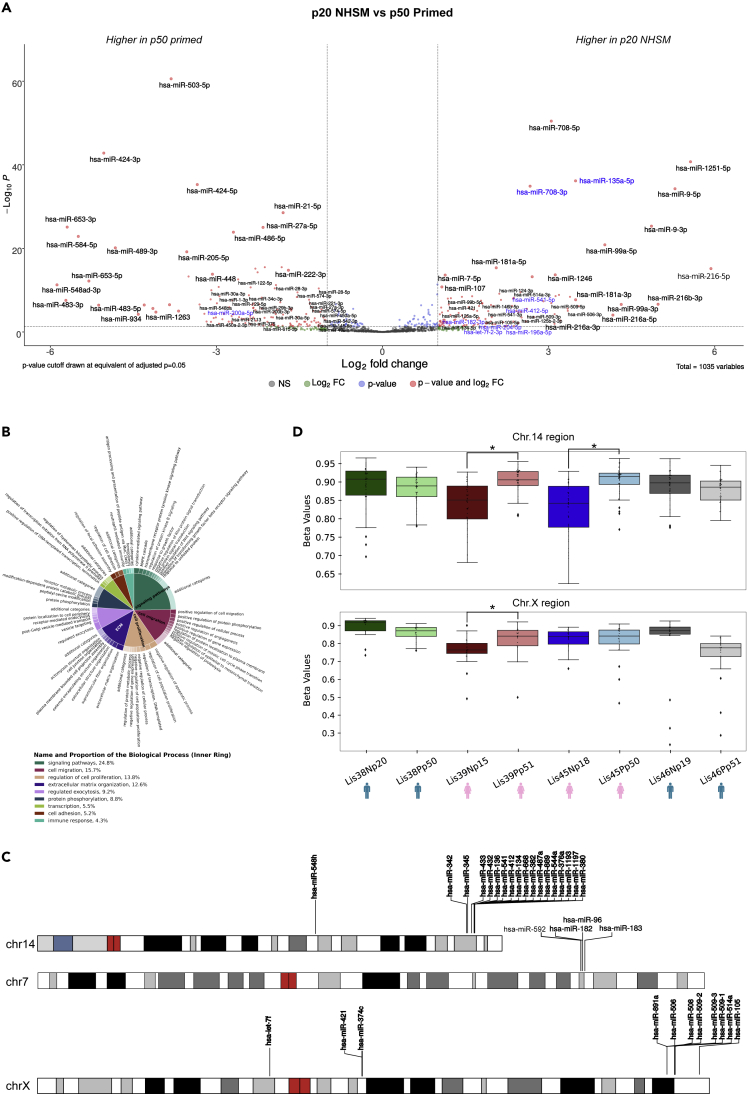
Figure 5.
miRNA differences between our NHSM and primed cultures
(A) Differential expression analysis between our NHSM and primed cultures. Volcano plot shows miRNAs significantly different (adj. p-value < 0.05, log2 fold change > |1|, basemean > 10) between our derivation timepoint samples (p20) and our late (p50) primed cultures. The dotted lines represent a log2 fold change of |1| and an adjusted p-value of 0.05. Grey dots are not significant miRNAs, blue dots are miRNAs that have an adj. p-value < 0.05 but not a log2 fold change > |1|, green dots have a log2 fold change > |1| but not an adj. p-value < 0.05, and red dots are those miRNAs considered significantly different by both metrics. miRNAs in blue font are those that were previously reported to be differentially expressed between NHSM and primed lines.
(B) Gene ontology (GO) enrichment analysis of downregulated genes (adj. p-value < 0.05, baseMean > 50) in the derivation timepoint that were also targeted by an miRNA upregulated in the derivation cultures. Revigo and CirGO were used to provide a non-redundant and biologically succinct visualization of GO terms with an adjusted p-value < 0.05.
(C) Genomic locations of miRNA clusters containing multiple miRNAs upregulated in derivation cultures (p20).
(D) Boxplots displaying methylation levels of the regions containing the chromosome 14 and chromosome X miRNA clusters in (C). Boxes span the top 75th percentile to bottom 25th percentile with the line in the middle of the boxes representing the median. Whiskers contain 95% of the data. “∗” represents a significant difference between cultures (p-value < 0.05 Mann–Whitney U).