Figure 3.
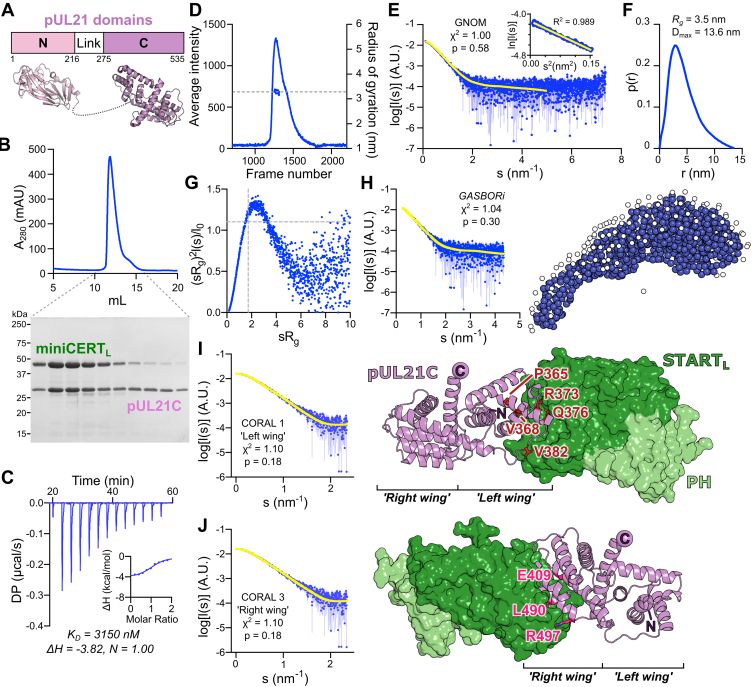
Solution structure of the H6-pUL21C–H6-miniCERTLheterodimer.A, schematic of pUL21, with N- and C-terminal domains joined by a highly dynamic linker (18). Crystal structures of domains (N, PDB ID: 4U4H (50); C, PDB ID: 5ED7 (51)) are shown. B, Superdex 75 10/300 SEC elution profile of the H6-pUL21C–H6-miniCERTL complex, preformed in the presence of 1.32-fold molar excess of H6-pUL21C. Dotted lines indicate fractions that were collected and subjected to SDS-PAGE analysis with Coomassie staining, revealing coelution of the two proteins. C, representative ITC titration curve of H6-pUL21C binding to H6-miniCERTL. Inset shows normalized binding curve with integrated changes in enthalpy (ΔH) as a function of molar ratio. The ranges of the vertical axes are identical to Figure 2F. The affinity (KD), ΔH, and stoichiometry (N) for the presented titration are displayed below. D, SEC elution profile (partial integrated scattered X-ray intensity versus data frame number) obtained during SEC–SAXS analysis of H6-pUL21C–H6-miniCERTL complex. Dashed line indicates the calculated radius of gyration (Rg) across the frames averaged for structural analyses. E, averaged SAXS profile of the H6-pUL21C–H6-miniCERTL complex. The reciprocal-space fit of the p(r) profile to the SAXS data is shown as a yellow line. χ2, fit quality; p, Correlation Map (CorMap) probability of systematic deviations between the model fit and the scattering data (92). Inset displays the Guinier plot (sRg < 1.3), which is linear as expected for an aggregate- and repulsion-free system. F, real-space distance distribution function, p(r), calculated from the SAXS profile. G, dimensionless Kratky plot of the SAXS data. Gray dotted lines indicate the expected maximum of the plot for a compact protein (sRg = √3, (sRg)2I(s)/I(0) = 3e−1). H, ab initio modeling of H6-miniCERTL–H6-pUL21C using GASBOR. Fit of the calculated scattering (yellow) to the SAXS profile is shown, as is a representative dummy-residue model (blue spheres) with modeled water beads of the hydration shell (white spheres). I and J, pseudoatomic models of the H6-miniCERTL–H6-pUL21C complex generated using CORAL. The fits of the computed scattering (yellow) to the H6-pUL21C–H6-miniCERTL SAXS profile (blue) are shown. High-quality fits are obtained with models where miniCERTL binds either the left or right “wings” of the dragonfly-like pUL21C domain (51). pUL21C is shown as a violet ribbon with “wings” and termini labeled, and miniCERTL as a green molecular surface (PH, light green; STARTL, dark green). For clarity, regions absent from the crystal structures that were modeled by CORAL are not displayed. Residues from the left and right “wings” of pUL21C that were selected for further investigation are shown as sticks with red (I) or pink (J) carbon atoms, respectively. An additional pseudoatomic model plus its fit to the H6-pUL21C–H6-miniCERTL SAXS profile is shown in Figure S3, where the fits presented in (I) and (J) are also reproduced for ease of comparison. DP, differential power; ITC, isothermal titration calorimetry; PDB, Protein Data Bank; PH, Pleckstrin homology; SAXS, small-angle X-ray scattering; SEC, size-exclusion chromatography; START, steroidogenic acute regulatory–related lipid transfer.