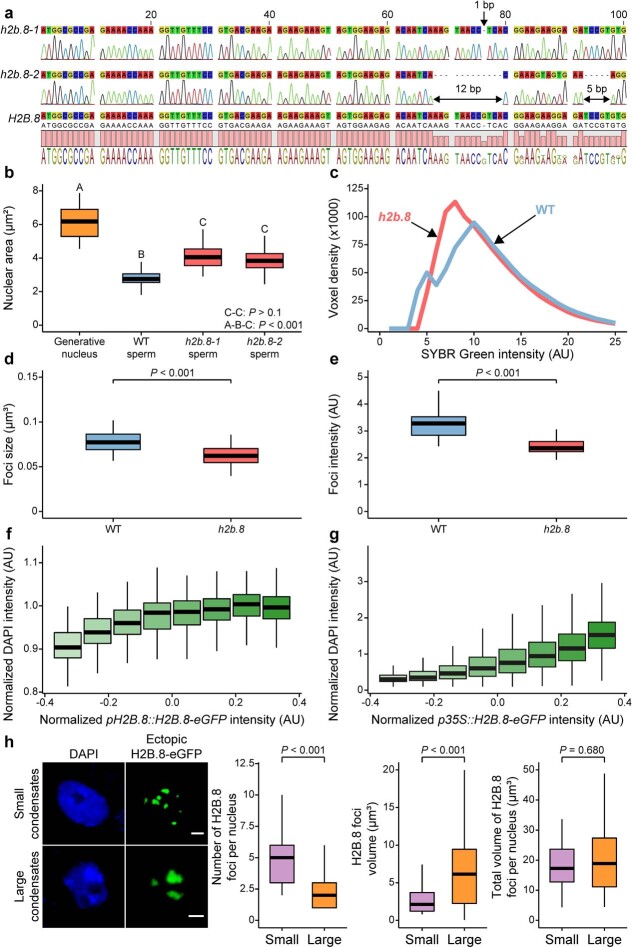
Extended Data Fig. 2. H2B.8 is required for sperm chromatin aggregation.
a, Alignment of h2b.8 CRISPR lines. h2b.8-1 has a single base deletion at 76 bp as indicated by an arrow, leading to a premature stop codon after 33 amino acids. h2b.8-2 has a 12 bp deletion after 67 bp and another of 5 bp after 92 bp, producing a truncated 54 amino acid protein. b, Nuclear sizes of the generative nucleus and sperm nuclei of indicated genotypes. P-values, one-sided ANOVA followed by individual two-sample Tukey tests, which adjust for multiple comparisons. Boxplots marked as A, B and C are significantly different between groups (P < 0.001) but not within the group (P > 0.1). n = 30 (generative nucleus), 80 (WT, h2b.8-1), and 77 (h2b.8-2) nuclei examined over two independent experiments. c, Density plot of individual SYBR Green-stained voxel intensities from wild-type (WT, blue) and h2b.8 (red) sperm nuclei. n = 30 nuclei for each. d, e, Boxplots indicating the volumes (d) and intensity (e) of SYBR Green foci in WT (blue) and h2b.8 (red) sperm. P-value, independent two-sample t-test. n = 30 nuclei for each. AU, arbitrary unit. f, g, Colocalization of DAPI and H2B.8-eGFP signals in sperm (f, pH2B.8::H2B.8-eGFP) and in H2B.8 ectopically expressed seedlings (g, p35S::H2B.8-eGFP). n = 37 and 31 nuclei for sperm and seedlings, respectively. h, Confocal images and quantification of H2B.8 condensates in p35S::H2B.8-eGFP seedlings. P-value, independent two-sample t-test. n = 30 and 71 for nuclei with small and large H2B.8 condensates, respectively, examined over two independent experiments. Scale bars, 2 μm.