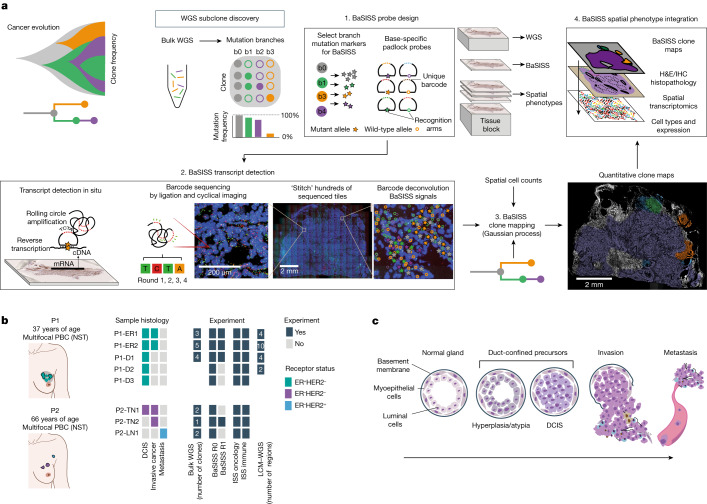
Fig. 1. The BaSISS workflow to generate cancer clone maps.
a, Following de novo mutation detection and subclone discovery in WGS data, the BaSISS workflow is performed as follows: (1) bespoke mutation-specific padlock probes are designed. (2) BaSISS transcripts are detected. To achieve this, BaSISS padlock probes hybridize to complementary DNA (cDNA) in situ. By virtue of a highly specific DNA ligase, only completely target-complementary padlock probes are ligated and form closed circles. Ligated probes are amplified through rolling circle amplification and their reader barcodes are detected in tissue space through sequencing by ligation with fluorophore-labelled interrogation probes and cyclical microscopy. (3) Mathematical modelling of BaSISS signals and the genotype of clones is then performed to derive clone maps. (4) Subsequent phenotype and microenvironment characterization of clones is then possible, by integrating clone fields with spatial datasets acquired from serial tissue sections. The BaSISS model and cell typing are described further in Extended Data Figs. 1 and 2. b, The two cases of multifocal primary breast cancer (PBC) used to develop the BaSISS approach. Coloured tiles report the histological features within each sample and the experiments performed. The number of clones identified by WGS and targeted by BaSISS are reported as white numerals. c, The traditional histological model of breast cancer progression. DCIS, ductal carcinoma in situ; H&E, haematoxylin and eosin; LN, lymph node; NST, invasive carcinoma of no special type; TME, tumour microenvironment.