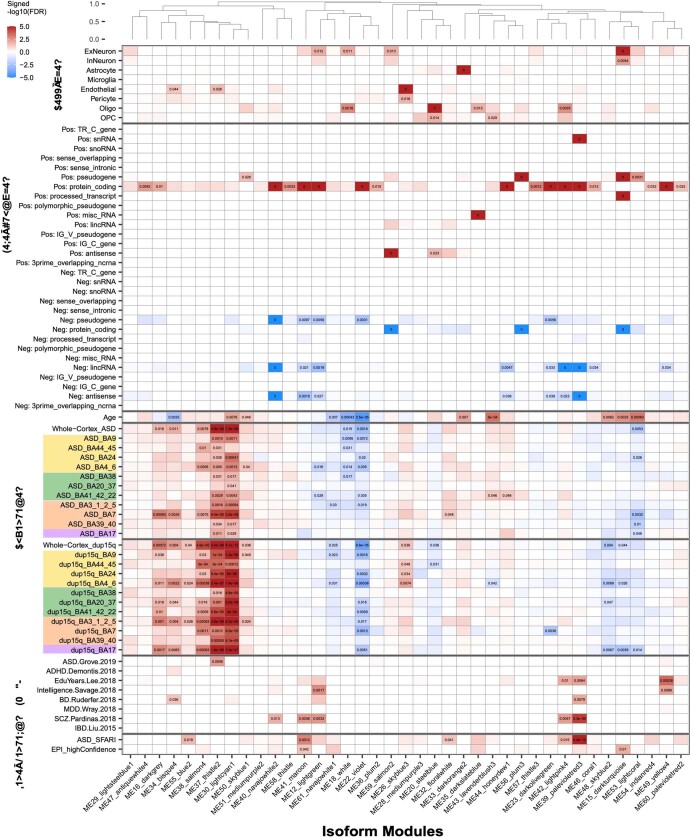
Extended Data Fig. 8. Transcript-level Co-Expression Network Analysis Module Associations.
Top: average-linkage hierarchical clustering of module eigengene biweight midcorrelations. Significant FDR corrected p-values (two sided) are indicated (FDR < 0.05; for GWAS, FDR < 0.1). For ASD, dup15q, and age covariates, FDR-corrected p-value from the linear mixed model testing the association of these covariates with module eigengenes is depicted. For the ASD and dup15q region-specific comparisons, cortical lobule colours are indicated (Fig. 1a). For gene biotypes, both positive and negative enrichment is shown (Fisher’s exact test; Methods). Positive enrichment is shown for cell-type, GWAS, and rare variant enrichments (Methods).