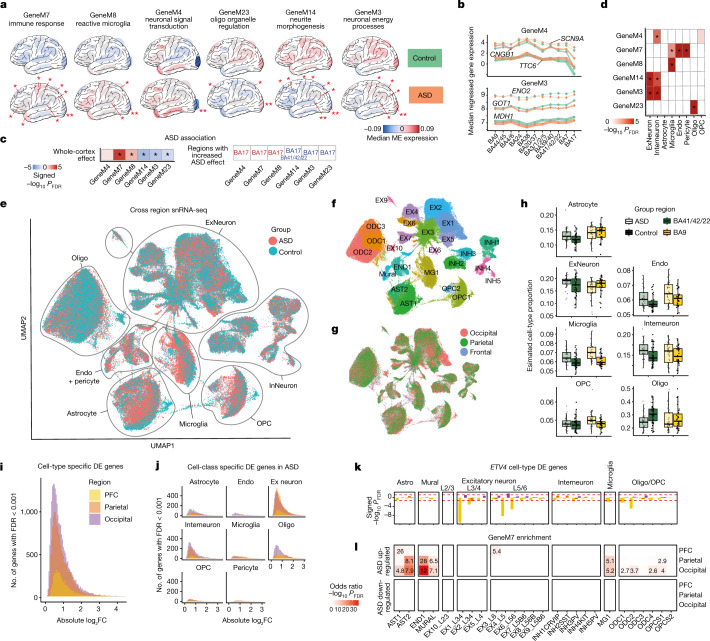
Fig. 4. Functional characterization of regionally variable transcriptomic dysregulation in ASD.
a, The top 6 most dysregulated modules with regionally variable patterns of dysregulation. The median of the module eigengene (ME), stratified by diagnosis, is depicted for each cortical region examined. *Significant region-specific dysregulation in ASD; **regions with a significantly increased magnitude of effect compared to the whole-cortex effect (Methods). b, Median regressed gene expression for the top three hub genes for GeneM4 (top) and GeneM3 (bottom). c, The whole-cortex ASD effect for modules depicted in a (*FDR < 0.05). Bottom, regions with a significantly increased magnitude of effect compared to the whole-cortex effect in a are listed. d, Cell-type enrichment for regionally variable modules. e, Uniform manifold approximation and projection for dimension reduction (UMAP) plots of snRNA-seq data from around 250,000 cells containing matched ASD and neurotypical control samples across frontal, parietal and occipital cortices, coloured by diagnosis. f,g, UMAP plots coloured by specific cell type (f) and cortical region of origin (g). h, Broad neural cell-type proportions deconvolved from matching bulk methylation array data. No FDR-significant cell proportion shifts were observed in ASD. i, Region-specific effect-size changes for cell-type-specific differentially expressed genes (FDR < 0.001), with BA17 again showing the greatest transcriptomic changes. j, Differentially expressed genes are shown for broad cell classes. k, ETV4 shows posterior-predominant downregulation across multiple cell types in ASD. l, The immune module GeneM7 shows posterior-predominant enrichment among cell-type-specific differentially expressed genes in ASD.