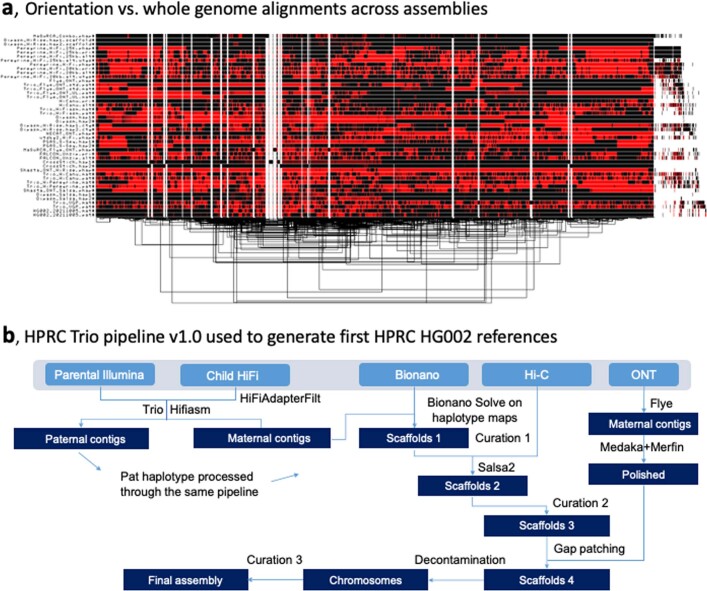
Extended Data Fig. 10. Pangenome alignment and generation of high-quality HPRC-HG002 v1.0 diploid assemblies.
a, Output of graph-based alignment of all chromosomes concatenated from all 45 HG002 assemblies (both haplotypes of diploid assemblies). Red vs Black, different orientations. Dendogram at bottom is a clustering of the alignments. b, HPRC v1.0 pipeline developed to produce the reference quality HPRC-HG002 v1.0 maternal and paternal assemblies. All steps shown are highlighted for the maternal data. The key steps of the pipeline are available in the Galaxy Server (https://assembly.usegalaxy.eu/) and best practices from this study at https://github.com/human-pangenomics/hpp_production_workflows/wiki/Assembly-Best-Practices.