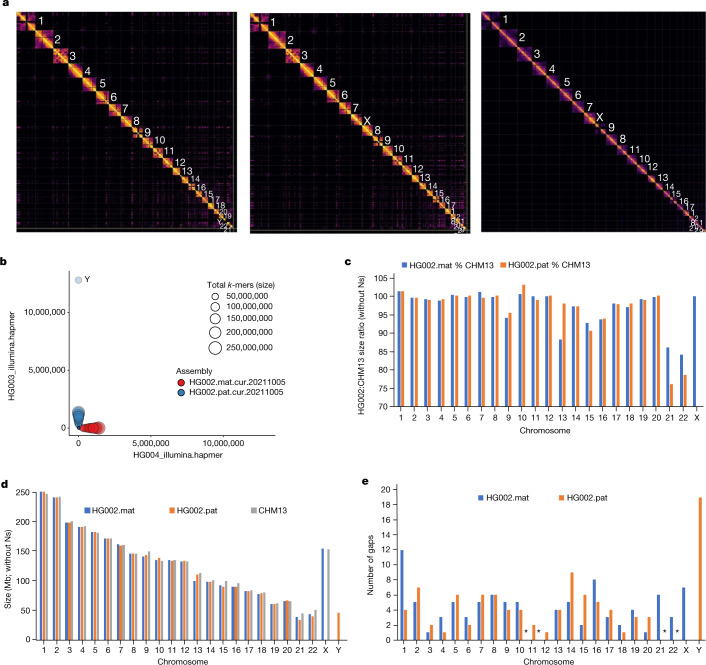
Fig. 3. Near-complete haplotype separation of scaffolds.
a, Hi-C contact maps to the final curated HPRC-HG002 paternal (left) and maternal (middle) assemblies in comparison to the CHM13 assembly (right). Values designate chromosome numbers, from largest to smallest size for each assembly. b, Blob plot using Illumina parental k-mers for the scaffolds of the HPRC-HG002 haplotypes. c, Percent size of HG002 diploid assembled chromosomes relative to CHM13 chromosomes, without including Ns. d, Comparison of absolute chromosome size values of all three assemblies, without including Ns. e, Number of remaining gaps in the chromosomes of each HG002 haplotype. Asterisks indicate assembled contigs with no gaps: maternal chromosomes 11 and 12, and assembled paternal chromosomes 21 and 22 without complete short arms.