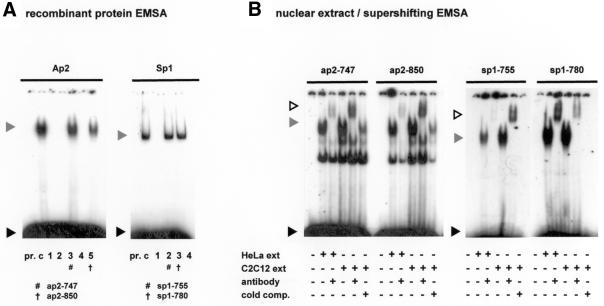
Figure 2.
Specific regions of the core utrophin promoter A region bind Ap2 and Sp1. (A) Double-stranded nucleotides spanning conserved binding sites of Ap2 and Sp1 were allocated a number corresponding to their location in the human promoter sequence (Fig. 1) and assayed with the appropriate recombinant transcription factor to determine binding ability. Formation of specific promoter A DNA–protein complexes are indicated by hash/cross marks. A grey arrowhead indicates the protein–DNA complex, with the black arrowhead indicating unbound DNA probe. Lane pr., consensus oligonucleotide only; lane c, control lane containing the consensus oligonucleotide and appropriate transcription factor. For Ap2: lane 1, ap2-709; lane 2, ap2-720; lane 3, ap2-747; lane 4, ap2-810; lane 5, ap2-850. For Sp1: lane 1, sp1-709; lane 2, sp1-755; lane 3, sp1-780; lane 4, sp1-850. (B) Formation of protein–DNA complexes using nuclear extract EMSA supershift analysis. Regions of interest were incubated with nuclear extract (30 µg HeLa and 15 µg C2C12) and antibodies (1.2 µg) as indicated. Cold comp. indicates the addition of a 100 or 500 molar excess of unlabelled probe for the Ap2 and Sp1 regions, respectively. Grey arrows indicate protein–DNA complexes that are supershifted in the presence of factor antibodies (white arrow). A black arrow indicates unbound labelled probe.