Fig. 1. Lineage-selective GATA4 chromatin occupancy in fetal heart.
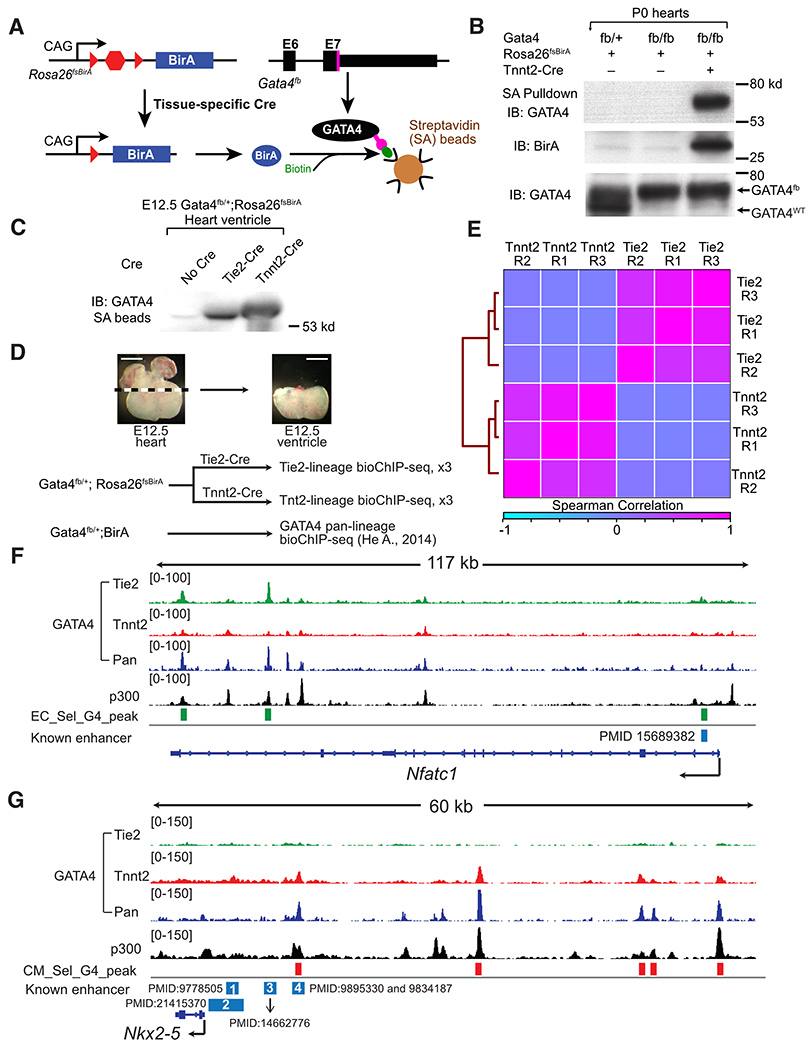
A. Schematic of Cre-directed in vivo GATA4 biotinylation. GATA4 with a C-terminal Flag-bio epitope tag is biotinylated by BirA, expressed in Cre-marked cells. B. Western blot demonstrating Cre-dependent GATA4 biotinylation and streptavidin (SA) pull down. P0 heart extract was incubated with SA beads. Input and SA-bound proteins were probed for GATA4 or BirA. C. GATA4 biotinylation in E12.5 CMs or ECs, directed by Tnnt2-Cre or Tie2-Cre. Heart lysate proteins bound to SA beads were probed with GATA4 antibody. D. Overview of GATA4 bioChIP-seq in CM and EC lineages. Pan-lineage GATA4 data was reported in ref. 11 Representative images show dissected tissues used for GATA4 bioChIP-seq. Bar, 500 μm. E. Correlation of lineage selective GATA4 bio-ChIP-seq data. Clustered heatmap displays the Spearman correlation across the union of peak regions. F,G. Representative genome browser views of lineage-selective GATA4 bioChIP-seq data at an ECC (Nfatc1) and a CM (Nkx2-5) gene. “Known enhancers” were validated by transient transgenesis in the indicated manuscripts. In F, the known enhancer is no longer active in ventricular ECCs by E12.5. In G, known enhancer 4, but not 1-3, are active in E12.5 heart. E12.5 heart ventricle p300 bioChIP-seq data was previously reported13.
