Figure 3.
Transcriptomic comparison of iHSCs, qHSCs, pHSCs, and fibroblasts
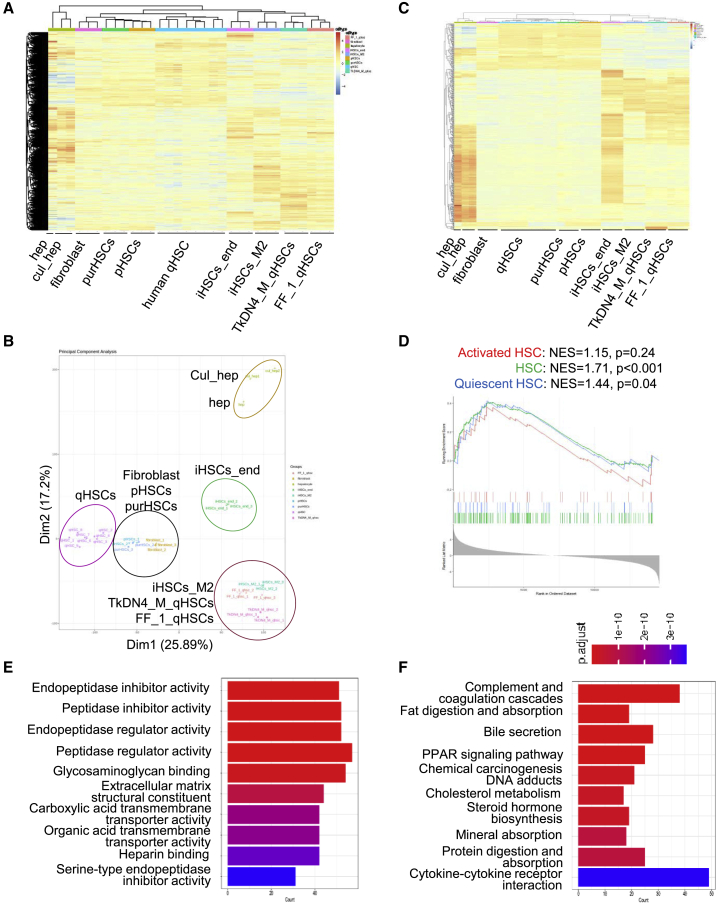
(A) Representative heatmap of transcriptomic profiles of endoderm-derived iHSCs (iHSCs_end, n = 3) and mesoderm-derived iHSCs (iHSCs_M2, n = 3), purchased primary HSCs (purHSCs, n = 3), primary HSCs isolated from liver tissue (pHSC, n = 3), quiescent HSCs (qHSCs, n = 8) (GSE141100), iPSC-derived qHSCs (TkDN4_M_qHSCs, n = 3, and FF_1_qHSCs, n = 3) (GSE155017), human hepatocytes (hep, n = 1) (GSE43984), cultured hepatocytes (cul_hep, n = 2, GSE98710), and fibroblasts (n = 3).
(B) PCA of the transcriptomic comparison of endoderm-derived iHSCs (iHSCs_end, n = 3) and mesoderm-derived iHSCs (iHSCs_M2, n = 3), purchased primary HSCs (purHSCs, n = 3), primary HSCs purified from liver tissue (pHSC, n = 3), quiescent HSCs (qHSCs, n = 8) (GSE141100), iPSC-derived qHSCs (TkDN4_M_qHSCs, n = 3, and FF_1_qHSCs, n = 3) (GSE155017), human hepatocytes (hep, n = 1) (GSE43984), cultured hepatocytes (cul_hep, n = 2, GSE98710), and fibroblasts (n = 3).
(C) Heatmap of the top 500 HSC genes expressed differently in endoderm-derived iHSCs (iHSCs_end, n = 3) and mesoderm-derived iHSCs (iHSCs_M2, n = 3), purchased primary HSCs (purHSCs, n = 3), primary HSCs purified from liver tissue (pHSC, n = 3), quiescent HSCs (qHSCs, n = 8) (GSE141100), iPSC-derived qHSCs (TkDN4_M_qHSCs, n = 3, and FF_1_qHSCs, n = 3) (GSE155017), human hepatocytes (hep, n = 1) (GSE43984), cultured hepatocytes (cul_hep, n = 2, GSE98710), and fibroblasts (n = 3).
(D) GSEA of the iHSC gene expression profile with qHSC and aHSC (GSE141100) gene signatures according to the phenotype described. NES, normalized enrichment score.
(E) GO analysis of 959 high-fold-change genes between iHSCs_end and TkDN4_M_qHSCs (GSE155017). The p values of the top 20 terms are indicated.
(F) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of 959 high-fold-change genes between endoderm-derived iHSCs (iHSCs_end) and other iPSCs derived qHSC-like cells from another laboratory (TkDNA4_M_qHSCs, GSE155017). The p values of enriched pathways are indicated. Data are represented as the mean ± SEM. See also Table S1.