| Impurity | Chemical name | CAS No. | Structure | QSAR ToolBox |
|---|---|---|---|---|
| – | Neohesperidine dihydrochalcone (E 959) | 20702‐77‐6 |
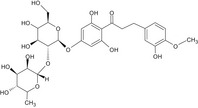
|
DNA binding OASIS: none DNA binding by OECD: Michael addition, P450‐mediated activation to quinone and quinone‐type chemicals (Hydroquinones) Carcinogenicity (gentox and nongentox): none DNA alerts for Ames, CA and MNT by OASIS: none In vitro mutagenicity (Ames) by ISS: none In vivo MN by ISS: Hacceptor‐path3‐Hacceptor (non‐covalent binding with DNA and/or proteins) Protein binding alerts for CA by OASIS: none |
| 1 | Phloroacetophenone neohesperidoside | – |
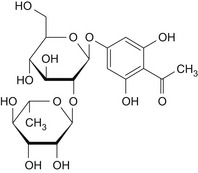
|
By read across with CAS 480‐66‐0; 1‐(2,4,6‐trihydroxyphenyl)ethenone: DNA binding OASIS: none DNA binding by OECD: none Carcinogenicity (gentox and nongentox): none DNA alerts for Ames, CA and MNT by OASIS: none In vitro mutagenicity (Ames) by ISS: none In vivo MN by ISS: Hacceptor‐path3‐Hacceptor (non‐covalent binding with DNA and/or proteins) Protein binding alerts for CA by OASIS: AN2 Michael addition to the quinoid‐type structures (Hydroxylated phenols) |
| 2 | Neodiosmin | 38665‐01‐9 |
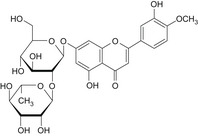
|
DNA binding OASIS: AN2 Michael‐type addition, quinoid structure; radical mechanism via ROS formation (Flavonoids) DNA binding by OECD: Michael addition, P450‐mediated activation to quinone and quinone‐type chemicals (Hydroquinones) Carcinogenicity (gentox and nongentox): none DNA alerts for Ames, CA and MNT by OASIS: none In vitro mutagenicity (Ames) by ISS: none In vivo MN by ISS: Hacceptor‐path3‐Hacceptor (non‐covalent binding with DNA and/or proteins) Protein binding alerts for CA by OASIS: Michael addition to the quinoid type structures, Schiff base formation, ROS generation (Arenecarbonyl compounds) |
| 3 | Neohesperidin | 13241‐33‐3 |
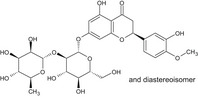
|
DNA binding OASIS: none DNA binding by OECD: Michael addition, P450‐mediated activation to quinone and quinone‐type chemicals (Hydroquinones) Carcinogenicity (gentox and nongentox): none DNA alerts for Ames, CA and MNT by OASIS: none In vitro mutagenicity (Ames) by ISS: none In vivo MN by ISS: Hacceptor‐path3‐Hacceptor (non‐covalent binding with DNA and/or proteins) (1,3‐dialkoxy‐benzene) Protein binding alerts for CA by OASIS: none |
| 4 | Naringin dihydrochalcone | 18916‐17‐1 |
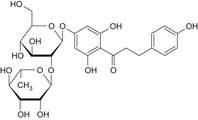
|
DNA binding OASIS: none DNA binding by OECD: Michael addition, P450‐mediated activation to quinone and quinone‐type chemicals (Alkyl phenols) Carcinogenicity (gentox and nongentox): none DNA alerts for Ames, CA and MNT by OASIS: none In vitro mutagenicity (Ames) by ISS: none In vivo MN by ISS: Hacceptor‐path3‐Hacceptor (non‐covalent binding with DNA and/or proteins) Protein binding alerts for CA by OASIS: none |
| 5 | Hesperidin dihydrochalcone | 35573‐79‐6 |
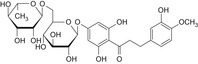
|
By read across from E959: DNA binding OASIS: none DNA binding by OECD: Michael addition, P450‐mediated activation to quinone and quinone‐type chemicals (Hydroquinones) Carcinogenicity (gentox and nongentox): none DNA alerts for Ames, CA and MNT by OASIS: none In vitro mutagenicity (Ames) by ISS: none In vivo MN by ISS: Hacceptor‐path3‐Hacceptor (non‐covalent binding with DNA and/or proteins) Protein binding alerts for CA by OASIS: none |
| 6 | Hesperetin dihydrochalcone 7′glucoside | – |
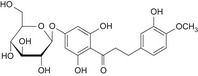
|
By read across from 35400‐60‐3: DNA binding OASIS: none DNA binding by OECD: Michael addition, P450‐mediated activation to quinone and quinone‐type chemicals (Hydroquinones) Carcinogenicity (gentox and nongentox): none DNA alerts for Ames, CA and MNT by OASIS: none In vitro mutagenicity (Ames) by ISS: none In vivo MN: Hacceptor‐path3‐Hacceptor (non‐covalent binding with DNA and/or proteins) Protein binding alerts for CA by OASIS: AN2, Michael addition to the quinoid‐type structure (hydroxylated phenols) |
| 7 | Hesperetin dihydrochalcone | 35400‐60‐3 |
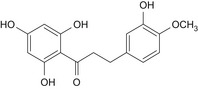
|
DNA binding OASIS: none DNA binding by OECD: Michael addition, P450‐mediated activation to quinone and quinone‐type chemicals (Hydroquinones) Carcinogenicity (gentox and nongentox): none DNA alerts for Ames, CA and MNT by OASIS: none In vitro mutagenicity (Ames) by ISS: none In vivo MN by ISS: Hacceptor‐path3‐Hacceptor (non‐covalent binding with DNA and/or proteins) Protein binding alerts for CA by OASIS: AN2, Michael addition to the quinoid‐type structure (hydroxylated phenols) |
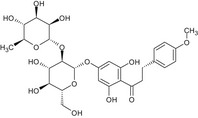
| 8 | Poncirin dihydrochalcone | – |
|
By read across from CAS 14941‐08‐3 (poncirin): DNA binding OASIS: none DNA binding by OECD: none Carcinogenicity (gentox and nongentox): none DNA alerts for Ames, CA and MNT by OASIS: none In vitro mutagenicity (Ames) by ISS: none In vivo MN by ISS: Hacceptor‐path3‐Hacceptor (non‐covalent binding with DNA and/or proteins) (1,3‐dialkoxy‐benzene) Protein binding alerts for CA by OASIS: none |

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.