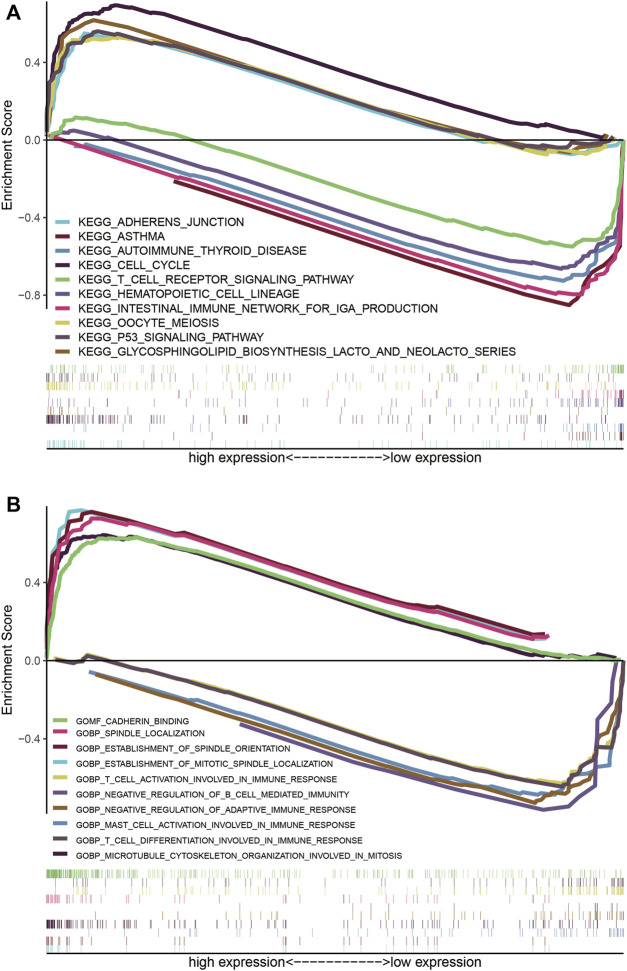
FIGURE 9.
LUADSenLncSig-based GSEA of different risk groups. (A) Based on the GSEA results, the KEGG genes were differentially enriched for senescence-related lncRNA expression. Five KEGG items, the cell cycle, p53 signaling pathway, oocyte meiosis, glycosphingolipid biosynthesis-lacto and neolacto series, and adherens junction were enriched in the high-risk group. Asthma, intestinal immune network for IgA production, hematopoietic cell lineage, autoimmune thyroid disease, and T cell receptor signaling pathway were enriched in the low-risk group based on the NES, NOM p-value, and FDR q-value (B) Differential enrichment of genes in GO with senescence-related lncRNAs. Five GO items, spindle localization, establishment of spindle orientation, establishment of mitotic spindle localization, cadherin binding, and microtubule cytoskeleton organization involved in mitosis showed a significant differential enrichment in the high expression phenotype. The other five GO terms, negative regulation of adaptive immune response, mast cell activation involved in immune response, T cell activation involved in immune response, T cell differentiation involved in immune response, and negative regulation of B cell mediated immunity, were found significantly enriched in the low expression phenotype based on the NES, NOM p-value, and FDR q-value. The reference files from the (MSigDB) were c2. cp.kegg.v7.4. symbols.gmt and c5. go.v7.4 symbols. gmt (http://software.broadinstitute.org/gsea/msigdb/index.jsp). LncRNAs, long non-coding RNAs; LUAD, lung adenocarcinoma; LUADSenLncSig, LUAD senescence lncRNA signature; GSEA, Gene Set Enrichment Analysis; KEGG, Kyoto Encyclopedia of Gene and Genome; NES, normalized enrichment score; NOM p-value, nominal p-value; FDR, false detection rate; GO, Gene Ontology; MSigDB, Molecular Signatures Database.