Figure EV1. Tetrahymena TtFer2, Tt00442310, and Tt00637180 are essential for mucocyst secretion (related to Fig 1).
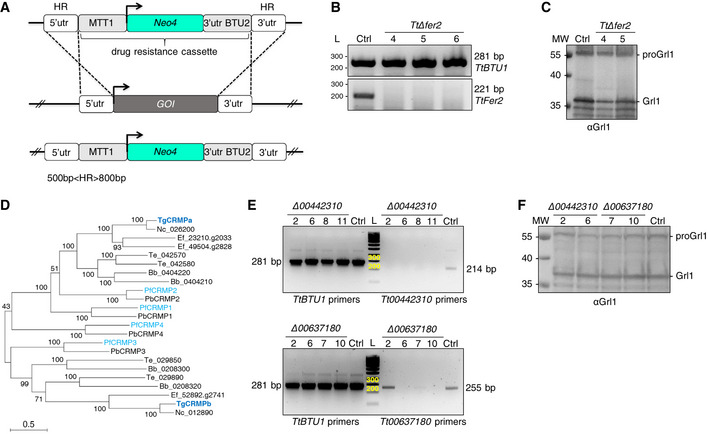
- Strategy for the macronuclear knockout of Tetrahymena thermophila genes of interest (GOI). A linearized construct carrying fragments (HR) homologous to the 5′ and 3′‐untranslated regions (UTR) of the GOI and flanking the drug resistance cassette were used to replace the GOI at the endogenous locus. The CdCl2‐inducible MTT1 promoter drives the expression of a paromomycin resistance gene (Neo4) used for selecting positive transformants.
- Disruption of the macronuclear copies of TtFer2 (ferlin 2; TTHERM_00886960) was assessed by RT–PCR. cDNA from wild‐type (Ctrl) and three clones of putative knockout cells (Δfer2) were PCR amplified with primers specific for TtBTU1 (β‐tubulin 1; upper panel) and TtFer2 (lower panel). The 221 bp products corresponding to transcripts from Fer2 are absent in the Δfer2 clones, indicating that all the wild‐type copies of TtFer2 were efficiently replaced with the Neo4 cassette. All samples showed wild‐type levels of BTU1 transcripts. L: DNA ladder (bp). Primers are listed in Table EV1.
- Western blot of whole‐cell lysates from wild‐type (Ctrl) and Δfer2 cells. In both, wild‐type and mutant extracts, anti‐Grl1 antibodies recognized the ~ 60 kDa precursor of the granule protein 1, proGrl1, and the processed form of Grl1, between 35 and 40 kDa, indicating non‐significant defects in proteolytic maturation. MW: molecular weight standards.
- Phylogeny depicting the relationships between Apicomplexa CRMPs. The maximum‐likelihood phylogenetic tree was obtained with the protein sequences of CRMP genes retrieved for the apicomplexans Toxoplasma gondii (TgCRMP), Plasmodium falciparum (PfCRMP), Plasmodium berghei (PbCRMP), Neospora caninum (Nc), Eimeria falciformis (Ef), Theileria equi (Te), and Babesia bigemina (Bb). Toxoplasma and P. falciparum CRMPs are highlighted in bold blue and light blue, respectively. Numbers at each node correspond to the bootstrap values. The scale bar represents the branch length.
- Disruption of the macronuclear copies of TTHERM_00442310 and TTHERM_00637180 was assessed by RT–PCR as in (B). Four clones for each putative knockout cell were tested. The 214 and 255 bp fragments corresponding to transcripts for TTHERM_00442310 and TTHERM_00637180, respectively, are absent in all Δ00442310 clones, and nearly undetectable in clones 6, 7, and 10 for Δ00637180, indicating the achievement of full knockout. Clones 2 and 6 for Δ00442310 and clones 7 and 10 for Δ00637180 were selected for further analysis. All samples show wild‐type levels of BTU1 transcripts. L: DNA ladder (bp). Primers are listed in Table EV1.
- Western blot of whole‐cell lysates from wild‐type (Ctrl), Δ00442310, and Δ00637180 cells. In both wild‐type and mutant extracts, anti‐Grl1 antibodies recognized processed Grl1 between 35 and 40 kDa and the precursor proGrl1 at ~ 60 kDa, indicating non‐significant defects in proteolytic maturation. MW: molecular weight standards.
Source data are available online for this figure.
