Figure EV4. TgCRMPa and TgCRMPb accumulate at the tip of the extruded conoid (related to Fig 5).
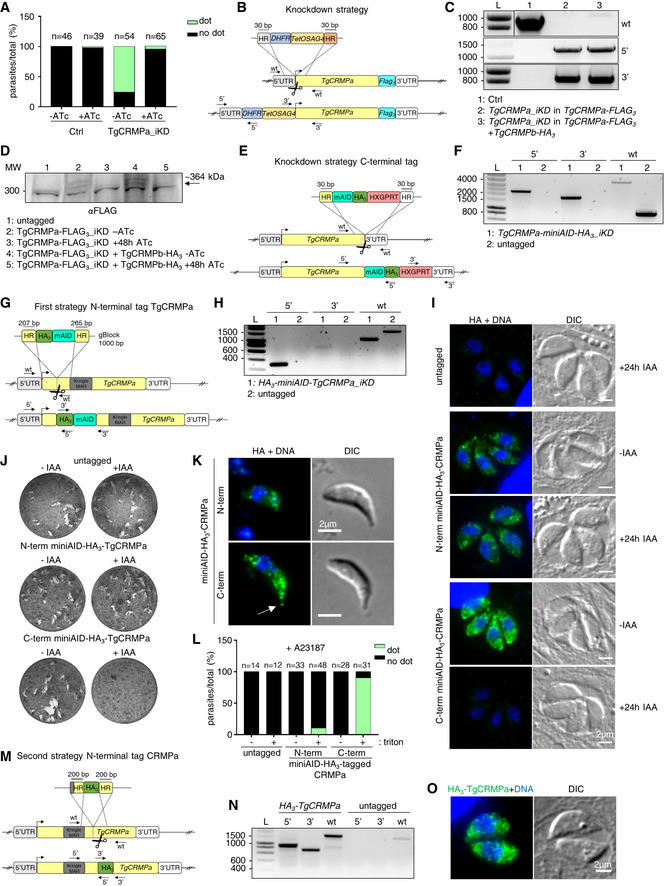
- Quantification of the dot pattern for TgCRMPa‐HA3 in TgCRMPa‐depleted (iKD) tachyzoites. TgCRMPa accumulation at the apical tip of extracellular parasites was measured upon incubation with host cell monolayers for 2 min to stimulate natural conoid extrusion. CRMPa signal at the apical dot disappeared after 48 h ATc treatment, indicating that the association with the tip of the extruded conoid was specific. No significant apical signal was detected for the control line (Ctrl), as in Fig 5B. Numbers are expressed as a percentage of parasites showing (dot) or lacking (no dot) the tip accumulation of TgCRMPa. The number of parasites (n) analyzed for each line is reported on the column tops.
- Strategy for the inducible depletion (iKD) of TgCRMPa‐FLAG3. The iKD lines were generated starting from the FLAG3‐tagged lines previously produced. In order to conditionally deplete the protein, the endogenous promoter of the TgCRMPa‐FLAG3 gene was replaced with an ATc‐regulableTetOSag4 promoter, preceded by the DHFR resistance cassette. The DNA fragment containing the cassette and promoter was PCR‐amplified from a donor vector with primers containing ~ 30‐bp‐long homology regions (HR) specific for TgCRMPa gene, and introduced upstream to the starting codon via CRISPR‐cas9 technology (scissors) and homologous recombination. The arrows indicate the binding sites of the primers used in (C).
- Integration of the DHFR cassette followed by the TetOSag4 promoter in the putative TgCRMPa‐FLAG3 and TgCRMPa‐FLAG3 + TgCRMPb‐FLAG3 iKD lines was tested by PCR as in Fig EV2C and D (upper panel). The fragments corresponding to the DHFR integration (5′) and TetOSag4 integration (3′) were detected exclusively in the putative iKD lines, while the wild‐type fragment (wt) was amplified only in the control line (Ctrl). L: DNA ladder (bp). Primers are listed in Table EV1.
- Whole‐cell lysates from untagged and TgCRMPa‐FLAG3_iKD and TgCRMPa‐FLAG3_iKD + TgCRMPb‐HA3 lines were immunoblotted with anti‐FLAG Abs to visualize tagged CRMPa in ATc‐treated and untreated samples. CRMPa disappeared upon 48 h ATc incubation in both lines. A ~ 300 kDa unspecific cross‐reactive band was observed in all samples. MW: molecular weight standards.
- Auxin‐degron strategy used for generating TgCRMPa‐miniAID‐HA3 strain. The integration of the tag and drug resistance cassette into the TgCRMPa locus is ensured by ~ 30‐bp‐long homology regions (HR) upon CRISPR‐Cas9 activity (scissors). The arrows indicate the binding sites of the primers used in (F).
- Integration of the miniAID‐HA3 and HXGPRT cassette at the TgCRMPa locus in the Tir‐1 line was tested by PCR as in Fig EV2A and B (upper panel). The fragments corresponding to the miniAID‐HA3 (5′) and HXGPRT cassette (3′) integration were detected exclusively in the putative iKD line, while the wild‐type fragment (wt) was amplified only in the untagged line. A ~ 4,000 bp fragment corresponding to the miniAID‐HA3 + HXGPRT cassette, and amplified with primers binding the wild‐type sequence, was detected in the iKD line. L: DNA ladder (bp). Primers are listed in Table EV1.
- Marker‐free strategy used for generating HA3‐miniAID‐TgCRMPa strain. The integration of the tag at the N‐terminus between residues Val69 and Leu70 (before the MAR/Kringle domain; Fig 5E) into the TgCRMPa locus is ensured by 207‐ and 265‐bp‐long homology regions (HR) flanking the tag in the synthetic gBlock, upon CRISPR‐Cas9 activity (scissors). The arrows indicate the binding sites of the primers used in (H).
- Integration of the miniAID‐HA3 at the N‐terminus of the TgCRMPa locus in the Tir‐1 parental line was tested by PCR. The fragments corresponding to the HA3 (5′) and the HA3‐miniAID (3′) integration were detected exclusively in the putative iKD line; the wild‐type fragment (wt) was amplified in the untagged line (~ 1,556 bp) and iKD line (~ 1,026 bp, tag minus introns). L: DNA ladder (bp). Primers are listed in Table EV1.
- Immunofluorescence images of untagged and N‐terminal and C‐terminal miniAID‐HA3‐TgCRMPa (iKD) intracellular tachyzoites. Parasites treated 24 h with IAA, as well as untreated (−IAA), were stained with anti‐HA Abs. The nuclei (DNA) are stained with Hoechst. Shown are single focal planes.
- Representative images of lytic plaques formation in HFF monolayers infected with IAA‐treated and untreated Tir‐1 control and N‐terminal and C‐terminal miniAID‐HA3‐TgCRMPa (iKD) lines.
- Immunofluorescence images of extracellular N‐terminal and C‐terminal miniAID‐HA3‐TgCRMPa tachyzoites. Parasites were incubated with ionophore A23187 to induce artificial conoid extrusion, and stained with anti‐HA Abs. TgCRMPa localization at the tip of the extruded conoid (arrow) is visible only in the C‐terminally miniAID‐HA3‐tagged TgCRMPa (lower panel). DNA is labeled by Hoechst. Single focal planes are shown. DIC: differential interference contrast.
- Quantification of the dot pattern for HA3‐miniAID‐TgCRMPa (N‐term) and TgCRMPa‐miniAID‐HA3 (C‐term) tachyzoites. TgCRMPa accumulation at the apical tip of extracellular parasites was measured upon incubation with ionophore A23187 to induce artificial conoid extrusion. Parasites were fixed and stained with anti‐HA Abs and with (+ triton) or without (− triton) permeabilization. CRMPa signal at the apical dot is absent in non‐permeabilized parasites, and it is robustly detected only in permeabilized parasites expressing C‐terminally miniAID‐HA3‐tagged TgCRMPa. No significant apical signal was detected for the control (untagged) or the N‐terminally miniAID‐HA3‐tagged TgCRMPa lines. Numbers are expressed as a percentage of parasites showing (dot) or lacking (no dot) the tip accumulation of TgCRMPa. The number of parasites (n) analyzed for each line is reported on the column tops.
- Marker‐free strategy used for generating HA3‐TgCRMPa strain. The integration of the tag at the N‐terminus between residues Thr600 and Asn601 (after the MAR/Kringle domain; Fig 5H) into the TgCRMPa locus is ensured by 200‐bp‐long homology regions (HR), flanking the tag in the synthetic gBlock upon CRISPR‐Cas9 activity (scissors). The arrows indicate the binding sites of the primers used in (N).
- Integration of the triple HA at the N‐terminus of the TgCRMPa locus was tested by PCR. The fragments corresponding to the 5′ and 3′ integration were detected exclusively in the putative HA3‐tagged line; and the wild‐type fragment (wt) was amplified in the untagged line (~ 1,424 bp) and tagged line (~ 1,550 bp, containing linker+HA3). L: DNA ladder (bp). Primers are listed in Table EV1.
- Immunofluorescence image of intracellular HA3‐TgCRMPa tachyzoites. Parasites were stained with anti‐HA Abs and DNA is labeled by Hoechst. Single focal planes are shown. DIC, differential interference contrast.
Source data are available online for this figure.
