Figure EV5. CRMPs and Nd6 co‐localize at the exocytic site in extracellular Toxoplasma gondii (related to Fig 6).
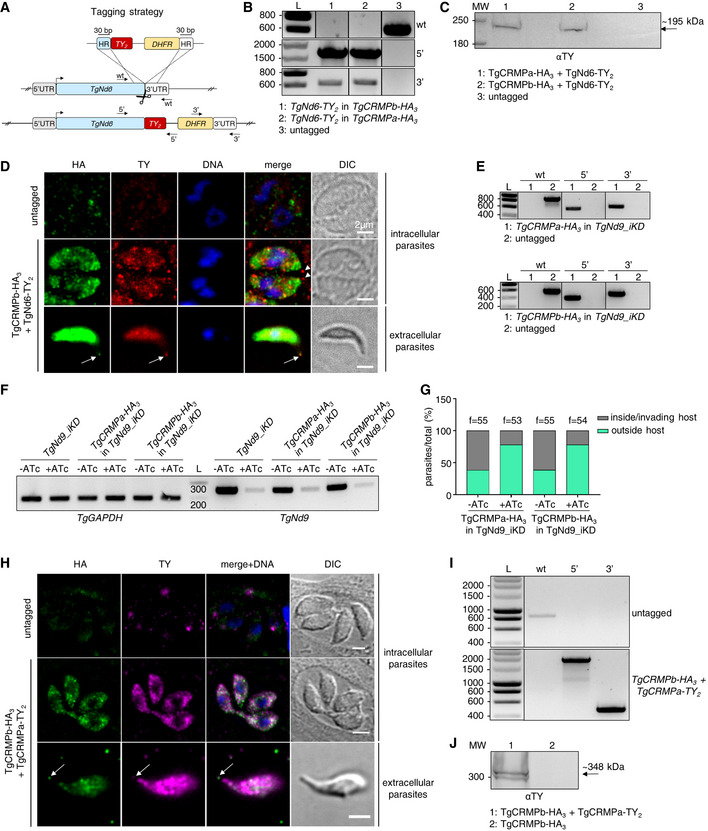
- Strategy for TY2 tagging of TgNd6 in TgCRMPa‐HA3 and TgCRMPb‐HA3 lines. To generate a C‐terminal TY2‐fusion of TgNd6, a DNA fragment was amplified from a donor vector containing the TY2 tag and the drug resistance cassette (DHFR). Primers to amplify the DNA fragment were designed to contain 30‐bp‐long stretches (HR) homologous to TgND6 regions flanking the insertion site for the epitope tag. Upon CRISPR‐cas9 cut (scissors), the PCR‐amplified DNA fragment efficiently recombines into the targeted endogenous locus. The arrows indicate the binding sites of the primers used in (B).
- Integration of the TY2 tag and DHFR cassette at the C‐terminus of TgND6 locus was tested by PCR. Genomic DNAs from an untagged line and clonal populations for TgNd6‐TY2 + TgCRMPa‐HA3 and TgNd6‐TY2 + TgCRMPb‐HA3 lines were amplified with primers binding to the 3′ C‐terminus and 3′UTR of TgNd6, and also in pairwise combination with primers binding the TY2 and DHFR sequences, respectively. The fragments corresponding to the TY2 tag (5′) and the resistance cassette (3′) were correctly amplified in the putative tagged lines, indicating that they were efficiently integrated at the TgNd6 locus. As expected, the wild‐type fragment for TgNd6 (wt) was detected only in the untagged line. L: DNA ladder (bp). Primers are listed in Table EV1.
- Whole‐cell lysates from untagged and TgNd6‐TY2 + TgCRMPa‐HA3 and TgNd6‐TY2 + TgCRMPb‐HA3 parasites were immunoblotted with anti‐TY Abs to detect tagged Nd6. A band around the expected size (~ 195 kDa) for TgNd6‐TY2 was observed exclusively for the tagged lines. MW: molecular weight standards.
- Immunofluorescence images of intracellular (upper and middle panels) and extracellular (lower panel) tachyzoites from untagged and TgCRMPb‐HA3 + TgNd6‐TY2 lines. Extracellular parasites were incubated with host cell monolayers for 2 min prior to fixation. Parasites were stained with anti‐HA and anti‐TY Abs to label CRMPb and Nd6, respectively. Nd6, but not CRMPb, accumulates at the tachyzoite apex in intracellular parasites (arrowheads), while both proteins localize at the tip of the extruded conoid in extracellular parasites (arrows). DNA is labeled by Hoechst. Single focal planes are shown. DIC, differential interference contrast.
- Integration of the HA3 tag and CAT cassette at the C‐terminus of TgCRMPa and TgCRMPb genes in TgNd9_iKD line was tested by PCR as in Fig EV2A and B. The fragments corresponding to the HA3 tag (5′) and the resistance cassette (3′) were correctly amplified in the putative tagged lines, indicating that they were efficiently integrated at the TgCRMPs loci. As expected, the wild‐type fragment of each gene (wt) was detected only in the untagged line. L: DNA ladder. Primers are listed in Table EV1.
- Depletion of TgNd9 transcripts was assessed by RT–PCR for the experiment shown in Fig 6D. Total RNAs from TgCRMPa‐HA3 and TgCRMPb‐HA3 expressed in TgNd9_iKD (minus epitope tag) parasites and parental line were subjected to reverse transcription and PCR amplified with primers binding TgNd9 transcripts. TgGAPDH was used as housekeeping gene. TgNd9 transcripts strongly decreased upon 72 h ATc treatment (+ATc). L: DNA ladder (L). Primers are listed in Table EV1.
- Depletion of TgNd9 proteins in the lines used for the experiment in Fig 6D was also assessed by quantifying the defect in the invasion of ATc‐treated TgNd9_iKD parasites expressing TgCRMPa‐HA3 and TgCRMPb‐HA3 versus untreated. The values are reported as percentages of the number of invading/intracellular and extracellular parasites over the total number of parasites. The number of fields (f) analyzed for each line is reported on the column tops.
- Immunofluorescence images of intracellular and extracellular tachyzoites from TgCRMPb‐HA3 + TgCRMPa‐TY2 line. Extracellular parasites were incubated with host cell monolayers for 2 min prior to fixation. Parasites were stained with anti‐HA and anti‐TY Abs to label CRMPb and CRMPa, respectively. Both proteins localize at the tip of the extruded conoid in extracellular parasites (arrows) and show partial overlap within the parasite cytosol. An untagged line was used to estimate the background noise. DNA is labeled by Hoechst. Single focal planes are shown. DIC, differential interference contrast.
- Integration of the TY2 tag and DHFR cassette at the C‐terminus of the TgCRMPa locus in TgCRMPb‐HA3 line was tested by PCR as described in (B) for TgNd6‐TY2. The fragments corresponding to the TY2 tag (5′) and the resistance cassette (3′) were correctly amplified in the putative tagged line, indicating that they were efficiently integrated at the TgCRMPa locus. As expected, the wild‐type fragment for TgCRMPa (wt) was detected only in the untagged line. L: DNA ladder (bp). Primers are listed in Table EV1.
- Whole‐cell lysates from TgCRMPb‐HA3 and TgCRMPb‐HA3 + TgCRMPa‐TY2 parasites were immunoblotted with anti‐TY Abs to detect tagged CRMPa. A band around the expected size (~ 348 kDa) for TgCRMPa‐TY2 together with the processed form were observed exclusively for the tagged line. MW: molecular weight standards.
Source data are available online for this figure.
