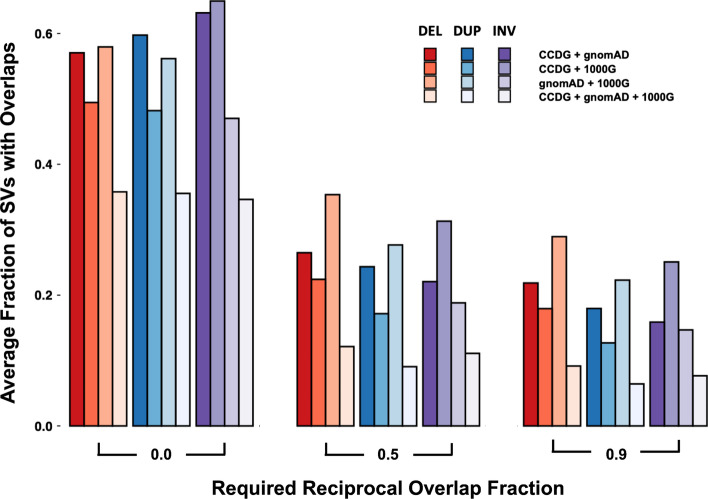
Fig. 1.
Matching SVs from different datasets based on shared SVTYPE and genomic overlaps. The average fraction of overlaps between deletions (DEL, in red), duplications (DUP, in blue), and inversions (INV, in purple) from CCDG, gnomAD, and 1000G are identified using varying amounts of required reciprocal overlap. Higher required reciprocal overlap fractions correspond to more exact genomic coordinate matches. Each dataset is compared to one another (CCDG + gnomAD, CCDG + 1000G, and gnomAD + 1000G) and overlaps with a different required reciprocal fraction are calculated. The fraction of total SVs found to have overlaps given the required reciprocal overlap fraction is found for each respective dataset and the average of these fractions is plotted. Finally, the average fraction of SVs found to have overlaps in all datasets (CCDG + gnomAD + 1000G) is found for each SVTYPE and at each required reciprocal overlap fraction