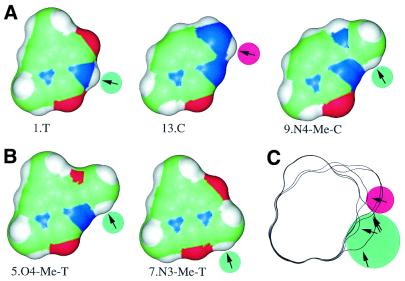
Figure 5.
Rescuing C binding affinity. Structures of bases and potential hydrogen bonds at position +7 are shown. Lower numbers correspond to stronger binding. (A) 13.C/A binds RepA more weakly than 1.T/A. This low binding can be rescued by substituting an N4 proton in C (13.C/A) with the methyl group in N4-Me-C (9.N4-Me-C/A). (B) Two more structures that have a C-H proton near the N3 atom also bind RepA well (5.O4-Me-T/A and 7.N3-Me-T/A). (C) Outlines of the structures in A and B were aligned. Arrows point in the direction from which RepA would have to approach to form hydrogen bonds with the protons (green circles). The hydrogen on N4 of 13.C is apparently pointing in the wrong direction or is too far from RepA to make a contact (arrow with red circle) since RepA approaches from the minor groove (bottom of the figure). To generate the figure, deoxyribose moieties were replaced by methyl groups and the structures were energy minimized for 1000 steps with the CV force field (Insight II, Molecular Simulations Inc., San Diego, CA). Connolly surfaces are shown (33).