Abstract
Upf1, Upf2, and Upf3, the central regulators of nonsense-mediated mRNA decay (NMD), appear to exercise their NMD functions while bound to elongating ribosomes, and evidence for this conclusion is particularly compelling for Upf1. Hence, we used selective profiling of yeast Upf1:ribosome association to define that step in greater detail, understand whether the nature of the mRNA being translated influences Upf1:80S interaction, and elucidate the functions of ribosome-associated Upf1. Our approach has allowed us to clarify the timing and specificity of Upf1 association with translating ribosomes, obtain evidence for a Upf1 mRNA surveillance function that precedes the activation of NMD, identify a unique ribosome state that generates 37–43 nt ribosome footprints whose accumulation is dependent on Upf1's ATPase activity, and demonstrate that a mutated form of Upf1 can interfere with normal translation termination and ribosome release. In addition, our results strongly support the existence of at least two distinct functional Upf1 complexes in the NMD pathway.
Keywords: selective ribosome profiling, Upf proteins, NMD substrates
INTRODUCTION
Nonsense-mediated mRNA decay (NMD) is a eukaryotic translation-dependent mRNA quality control pathway whose central regulators are the three Upf proteins, Upf1, Upf2, and Upf3 (He and Jacobson 2015; Kurosaki et al. 2019). NMD is initiated in response to atypical translation termination events, for example, when an elongating ribosome encounters a termination codon that occurs prematurely within an open reading frame (ORF) or in a context that otherwise renders termination inefficient (Amrani et al. 2004; Wu et al. 2020). Although NMD's existence has been known for decades (Losson and Lacroute 1979; Leeds et al. 1991; He et al. 1993; Maquat 2000), and the pathway has been studied extensively (He and Jacobson 2015; Kurosaki et al. 2019), the precise mechanism by which the Upf proteins recognize a ribosome undergoing an atypical termination event and respond to it by triggering accelerated degradation of the associated mRNA remains unknown.
Several observations have indicated that the NMD functions of the yeast Upf proteins are exercised while these factors are bound to ribosomes, and the evidence for this conclusion is particularly compelling for Upf1. This includes experiments demonstrating: (i) cosedimentation of Upf1 with polyribosomes (Atkin et al. 1997; Mangus and Jacobson 1999; Kashima et al. 2006), (ii) retained association of Upf1 with ribosomes when cytoplasmic extracts are digested with RNase A (Atkin et al. 1995) or micrococcal nuclease (Mangus and Jacobson 1999), (iii) substantial resistance of Upf1:ribosome association to prior treatment with high salt (Mangus and Jacobson 1999; Ghosh et al. 2010; Min et al. 2013; Celik 2017), (iv) two-hybrid interaction of Upf1 with the 40S ribosomal subunit protein Rps26 and elimination of this interaction by specific Upf1 CH domain C62Y and C84S mutations (Min et al. 2013), (v) interaction of Upf1 with the release factors (Czaplinski et al. 1998; Wang et al. 2001; Kashima et al. 2006; Ivanov et al. 2008), and (vi) a requirement for Upf1 in post-termination ribosome reutilization (Ghosh et al. 2010). As in yeast, Upf1 migrates with polyribosomal sucrose gradient fractions in mammalian cell extracts (Nott et al. 2004; Lopez-Perrote et al. 2016; Kurosaki et al. 2018; Yoshikawa et al. 2018). However, human Upf1 appears to bind mRNAs and be displaced from their coding regions to their 3′-UTRs by elongating ribosomes (Hogg and Goff 2010; Hurt et al. 2013; Kurosaki and Maquat 2013; Zund et al. 2013). The latter observations may reflect the involvement of Upf1 in numerous mammalian mRNA decay pathways other than NMD (Kim and Maquat 2019) or imply that Upf1 may respond in trans to an NMD-activating event during translation termination, possibly as part of a “two-factor authentication” process (Boehm et al. 2021).
In yeast, NMD substrates are generally thought to be decapped without prior deadenylation and to be subsequently degraded by the 5′ to 3′ exonuclease, Xrn1 (Muhlrad and Parker 1994). Recently, we demonstrated that Upf1 acts as a decapping activator for yeast NMD, controlling both substrate specificity and activation of the decapping enzyme by binding to two specific regulatory motifs in the Dcp2 carboxy-terminal domain (He et al. 2022). Significantly, the same study demonstrated that decapping is not rate-limiting for NMD substrates and that the major function for Upf1 in NMD is imparted at a step upstream of mRNA decapping. These observations indicate that there are substantial unknown aspects of Upf1 association with mRNAs or prematurely terminating ribosomes and the consequences of such association for determining a transcript's eligibility for NMD and/or for committing it to the NMD pathway.
Regardless of where Upf1 and the other Upf factors reside while awaiting their activation and involvement in mRNA decay, it appears that the steps in which these proteins are committed to NMD functions are likely to be localized to ribosomes (He and Jacobson 2015; Kurosaki et al. 2019). Hence, we used selective ribosome profiling (Becker et al. 2013) of yeast Upf1 to define the step and the nature of the mRNA being translated on Upf1:80S interaction, and to elucidate the functions of ribosome-associated Upf1. Our approach has allowed us to clarify the timing and specificity of Upf1 association with translating ribosomes, obtain evidence for a Upf1 mRNA surveillance function that precedes the activation of NMD, identify a unique ribosome state that generates 37–43 nt footprints whose accumulation is dependent on Upf1's ATPase activity, and demonstrate that a mutated form of Upf1 can interfere with normal translation termination.
RESULTS
Rationale for the methodological approaches
To characterize Upf1 association with polyribosomes, we carried out selective ribosome profiling analyses (Becker et al. 2013) of yeast cells expressing FLAG-tagged alleles of UPF1. To avoid recovery of Upf1 associated with extra-ribosomal complexes (Atkin et al. 1995; Mangus and Jacobson 1999), FLAG-based immunopurification (IP) was applied to RNase I-digested ribosomes that had been pelleted through a 1M sucrose cushion, a procedure yielding relatively pure 80S ribosome preparations (Supplemental Fig. S1). To address possible methodological or biological variables arising from tag location, UPF1 constructs with amino- or carboxy-terminal FLAG epitopes were examined. Although several protocols (Becker et al. 2013; Doring et al. 2017; Wagner et al. 2020) recommend that selective ribosome profiling be carried out with carboxy-terminally tagged proteins to preclude sequencing reads generated by immunopurification of ribosomes with a nascent amino-terminally tagged protein, we simply eliminated all Upf1-specific reads bioinformatically (Supplemental Table S1). To increase the possibility of characterizing steps in NMD upstream of the actual activation of mRNA decay, we examined cells with or without full NMD activity, including cells harboring upf mutations and cells that were briefly treated or not treated with cycloheximide (CHX) during harvesting and lysis. CHX inhibits translation elongation and CHX treatment has been shown to inhibit NMD in vivo (Zhang et al. 1997) and to block the formation of toeprints specific to premature termination codons in vitro (Amrani et al. 2004).
Pilot experiments in which FLAG-tagged Upf1 was expressed from centromeric plasmids did not yield sufficient immunopurified ribosomes for construction of substantive ribosome profiling libraries. Hence, episomally expressed UPF1 genes were used for all experiments. The increased expression inherent to such constructs raises the possibility that results obtained with their use might not reflect bona fide Upf1 functions or distribution across an mRNA coding region. However, several results mitigate this concern: (i) in vitro incubation of yeast Upf1 and 80S ribosomes at Upf1:80S ratios considerably higher than those achieved by in vivo episomal expression nevertheless yielded highly specific Upf1:80S interaction (Schuller et al. 2018), (ii) the distribution of WT Upf1-FLAG across different polysome fractions was comparable when the protein was expressed from centromeric or episomal vectors in WT UPF2 strains, even when free Upf1 was increased (Supplemental Fig. S2, middle, fractions 11 and 12, episomal Upf1-FLAG), whereas increased migration of Upf1-FLAG into heavier polysomes was observed when UPF2 was deleted, and (iii) episomal expression of UPF1 displayed a similar NMD phenotype compared to centromeric UPF1, causing only slight inhibition of NMD relative to endogenous UPF1 (Supplemental Fig. S3). Consistent with the latter northern blotting results, our RNA-seq analyses showed that episomally expressed UPF1 manifested negligible dominant-negative activity, that is, yeast NMD substrates (Celik et al. 2017) showed only small increases in abundance as compared to the substantial increases in abundance seen in upf1DE572AA and upf2Δ strains in which NMD is inactivated (Supplemental Fig. S4). Cells harboring episomally expressed UPF1 thus appear to maintain near normal NMD function, a conclusion further supported by additional experiments described below, including those in which we: (i) episomally expressed all three Upf proteins simultaneously, (ii) used upf1 or upf2 mutations to determine if a particular result depended on Upf1 or NMD function, and (iii) tested whether Upf1:ribosome complexes manifested stoichiometric relationships between Upf1 and ribosomal proteins.
Purification of FLAG-Upf1- and Upf1-FLAG-associated ribosomes yields stoichiometric recovery of ribosomal proteins
Confirmation of the specificity of recovery of Upf1-associated ribosomes followed from mass spectrometry and intensity-based absolute quantification of protein abundance (iBAQ) (Schwanhausser et al. 2011, 2013) analyses of ribosomes from two biological replicates of FLAG-UPF1 strains or three biological replicates of UPF1-FLAG strains harvested with or without addition of CHX. The normalized iBAQ values for the total and immunopurified samples were compared to each other in scatter plots and volcano plots (Fig. 1; Supplemental Tables S2, S3). Using ribosomes from cells untreated or briefly treated with CHX during cell harvesting and lysis, immunopurification resulted in substantial enrichment of FLAG-tagged Upf1 (Fig. 1B, red dots) such that, in all cases, its increased abundance after immunopurification approximated stoichiometry with the ribosomal proteins recovered in the samples (Fig. 1A, blue dots, and Supplemental Fig. S5), a result consistent with copurification of Upf1 with ribosomes. Specificity of the FLAG immunopurification procedure was demonstrated by greatly reduced recovery of ribosomes after immunopurification (Supplemental Fig. S6, α-Rps6), and the lack of increased Upf1: ribosomal protein stoichiometry, when the same procedure was applied to ribosomes from control cells expressing 6XHis-tagged UPF1 (Fig. 1A; Supplemental Fig. S5). The stoichiometry of Upf1:ribosomal proteins after immunopurification of ribosomes from cells expressing FLAG-UPF1 treated or not treated with CHX at the time of harvesting was similar (Fig. 1A, red dots, Y-axis values). Likewise, Upf1 derived from the carboxy-terminally FLAG-tagged UPF1 allele expressed in different genetic backgrounds was also recovered stoichiometrically with ribosomal proteins, indicating that tag location, deletion of UPF2, and the ATPase-inactivating upf1DE572AA mutation did not affect Upf1:ribosome association or its recovery. As expected for ribosomes subjected to RNase I digestion, immunopurification of Upf1-associated ribosomes did not lead to uniform recovery of all ribosomal proteins. Among those that are depleted in most IP samples are Rpp2A and Rpp2B (Fig. 1A,B), components of the ribosomal stalk (Hanson et al. 2004).
FIGURE 1.
Stoichiometric recovery of FLAG-tagged Upf1 with ribosomal proteins. Input (Total) and immunopurified ribosomes (IP) from different lysates were analyzed by mass spectrometry. Intensity-based absolute quantification (iBAQ) was performed and normalized across biosamples. (A) Average iBAQ of biological replicates, log10-transformed, of proteins identified from IP were plotted against those from the Total ribosomes. Proteins exclusively identified in either Total or IP are plotted on the x- or y-axis, respectively. (B) Differential abundance analysis was performed for proteins detected in both IP and Total samples using R package limma (Smyth 2004; Kammers et al. 2015). Negative log10 P-values adjusted by Benjamini–Hochberg method were plotted against log2 fold change in protein abundance in IP over Total. Gray vertical dashed line indicates log2 fold change of zero (no change). Positive log2 fold change (right of vertical line) are proteins enriched in IP. Negative log2 fold change (left of vertical line) are proteins depleted in IP. Gray horizontal dashed line indicates the cutoff of adjusted P-value at 0.05; proteins above this cutoff have significant changes. Two biological replicates of FLAG-UPF1 strains, three biological replicates of UPF1-FLAG strains, and one sample of the negative control experiment (6XHis-UPF1) were analyzed.
With the exception of the FLAG-tagged UPF1 gene, the plasmid selective markers, and the gene disruption cassette selective markers in upf1Δ and upf2Δ strains, all genes in these strains were present at their normal copy numbers. Thus, it was not surprising that immunopurification of FLAG-Upf1 or Upf1-FLAG did not lead to corecovery of the interacting Upf factors, Upf2 and Upf3. However, detectable levels of Upf2 and Upf3 were recovered after immunopurification of ribosomes from FLAG-UPF1 cells in which both untagged UPF2 and UPF3 were also expressed from episomal vectors (Fig. 1A, FLAG-UPF1 UPF2/3 EE +CHX), demonstrating that their lack of detection in other samples was due to lower relative abundance of endogenous Upf2 and Upf3 and not the consequence of loss during the immunopurification procedure. Immunopurification of ribosomes from the FLAG-UPF1 UPF2/3 EE +CHX cells also led to corecovery of the release factor eRF1 and enrichment for the mRNA decapping factors Edc3 and Dcp2 (Fig. 1A), factors that were not recovered in any of the other IP samples. However, many other proteins including translation initiation and elongation factors, and proteins unrelated to translation, mRNA decay, or protein folding were also enriched only in samples from this strain (Fig. 1B), so it is uncertain whether corecovery of release and decapping factors was specific to the presence of Upf2 and Upf3 on the ribosomes or conversely, whether simultaneous increased expression of all three Upf factors resulted in nonspecific recovery of these and other proteins. Having demonstrated the specificity of our immunopurification procedure, subsequent experiments used ribosome profiling and RNA-seq analyses of cells expressing FLAG-tagged Upf1 episomally.
Upf1 association with 80S ribosomes in CHX-treated cells promotes the formation of atypical ribosome-protected mRNA fragments
Ribo-Seq libraries were prepared from both immunopurified ribosomes and the respective prepurification total ribosomes derived from the complete set of strains expressing FLAG-tagged Upf1, as well as from wild-type cells harboring an empty vector (EV) control. We first examined the nature of the ribosome protected fragments recovered in all libraries. Analyses of ribosome protected footprint length distribution in FLAG-UPF1 libraries from cells without CHX treatment showed that footprints of ∼20–23 nt in length (“small” size, hereafter denoted as “S”) were predominant, followed by ∼27–32 nt footprints (“medium” size, hereafter denoted as “M”; Fig. 2A, green lines). These previously detected (Wu et al. 2019) footprint sizes were also present in control cells lacking FLAG-tagged UPF1 and in samples with or without prior immunopurification of Upf1-associated ribosomes. In the presence of CHX, the predominant footprint sizes from total or immunopurified ribosomes were the M size class; this is expected since CHX is known to freeze ribosomes with occupied A sites, yielding footprints ∼28 nt in length (Fig. 2A, red lines; Lareau et al. 2014; Wu et al. 2019). Notably, ribosomes from CHX-treated strains expressing FLAG-UPF1 yielded an atypical footprint size class of ∼37–43 nt (“large” size, hereafter denoted as “L”) (Fig. 2A, red solid lines). The L footprints represent translating ribosomes (i.e., they are neither RNA binding complexes nor scanning ribosomes), as do the typical S and M footprints, because they exhibited the 3-nt periodicity expected of translating ribosomes (Supplemental Fig. S7). The ratio of L:M footprints was increased in the FLAG-UPF1 IP libraries compared to total libraries (Fig. 2A, red solid lines), and the L footprints were not detectable in libraries prepared from cells without CHX treatment (Fig. 2A, green lines).
FIGURE 2.
Distribution of footprint length for each strain and cycloheximide treatment condition. Fractions of each footprint length (nt) were calculated and averaged among replicates. Green and red lines indicate libraries prepared in the absence and presence of cycloheximide, respectively. Dashed and solid lines represent total and immunopurified (IP) ribosomes, respectively. Gray shaded areas highlight different classes of footprint size: from left to right, 20–23 nt = small (S), 27–32 nt = medium (M), and 37–43 nt = large (L). (A) Data from libraries prepared from two biological replicates of amino-terminally FLAG-tagged strains and their WT + EV control. (B) Data from libraries prepared from three biological replicates of carboxy-terminally FLAG-tagged strains and their WT + EV control.
We considered the possibility that the atypical L footprints might be a consequence of nonstoichiometric expression of UPF2 or UPF3 relative to UPF1 in cells expressing episomal FLAG-UPF1, but concurrent episomal expression of UPF2, UPF3, and FLAG-UPF1 still yielded L footprints from immunopurified ribosomes (Fig. 2A, bottom panel). Likewise, we considered the possibility that the L footprints were caused by the 5′-FLAG epitope on Upf1, but analysis of libraries generated from cells expressing UPF1-FLAG again showed recovery of the L footprints in immunopurified ribosomes (Fig. 2B). Therefore, this unique class of footprints is most likely specific to Upf1 association with the 80S ribosome.
Upf1 function but not full activity of the NMD pathway is required for formation of atypical footprints by Upf1-associated ribosomes
To understand the origin of the L footprints, we determined whether their formation required Upf1 function, as well as function of the entire NMD pathway. Total and immunopurified ribosomes from CHX-treated upf1Δ strains harboring the upf1DE572AA-FLAG allele, or UPF2 or upf2Δ strains harboring the UPF1-FLAG allele, were subjected to ribosome profiling and analysis of footprint length distribution (Fig. 2B). The L footprints were undetectable in any libraries from total ribosomes but were recovered in libraries prepared from immunopurified ribosomes isolated from UPF2 or upf2Δ cells expressing UPF1-FLAG. The results indicate that full functionality of the NMD pathway is not required to form L footprints and that the position of the FLAG epitope does not influence L footprint formation. However, the L footprints were undetectable in libraries prepared from the upf1DE572AA-FLAG strain (Fig. 2B), indicating that full Upf1 function is required for their formation. Upf1-DE572AA is present on polysomes (Supplemental Fig. S2) and is able to interact with Rps26 in a two-hybrid assay (Min et al. 2013); therefore, the loss of the L footprints in cells expressing upf1DE572AA-FLAG is likely due to the inability of this mutant Upf1 to promote a ribosome-associated function that requires Upf1's ATP hydrolysis activity.
L footprints are generated by additional protection of mRNA 5′ to the normal ribosome-protected segment
The recovery of L footprints from CHX-treated cells expressing FLAG-UPF1 or UPF1-FLAG (Fig. 2A,B) suggests that a fraction of Upf1 is bound to ribosomes in a configuration that inhibits RNase I digestion of mRNA that usually occurs at the edge of the ribosome during library preparation. Mapping of the 5′ and 3′ ends of the M and L footprints recovered from Upf1-associated ribosomes using the start and stop codons as reference points showed that the 3′ ends but not the 5′ ends of M and L footprints are aligned at the same nucleotide location (Fig. 3). Thus, the size difference between M and L footprints is entirely attributable to an extension on the 5′ side of the normal ribosome-protected fragment. The 5′ ends of the typical M footprints are ∼12–13 nt upstream of the reference codon (start or stop codon), while the 5′ ends of the atypical L footprints are ∼23–25 nt upstream of the reference codon, regardless of the amino- or carboxy-terminal placement of the FLAG epitope, confirming that the extra 10–13 nt that extend nuclease protection do so on the 5′ side of the fragment. This corresponds to the region of the mRNA near the exit channel of the ribosome, the same region in which Upf1:80S interaction was observed in vitro (Schuller et al. 2018).
FIGURE 3.
Atypically large footprints are attributable to a 5′ extension of the mRNA region protected by the ribosome. Mapping of the 5′ and 3′ ends of footprint lengths 25–45 nt relative to the start and stop codons (red dashed lines) from IP libraries of FLAG-UPF1, UPF1-FLAG, and UPF1-FLAG/upf2Δ (+CHX). For each footprint length, footprint count at each nucleotide position relative to the start or stop reference codons were normalized to the total footprint count in the library, and these normalized counts from biological replicates were averaged. Different footprint lengths align at the 3′ ends but diverge at the 5′ ends. L and M fragment size classes are as indicated for each sample. Lines at the bottom of the figure indicate the approximate 5′ and 3′ limits of the L and M fragments over the start and stop reference codons.
L footprints reflect an early phase of Upf1 association with polyribosomes
Metagene analyses comparing the distributions of S, M, and L ribosome-protected fragments across normalized coding regions from total and immunopurified ribosomes from FLAG-UPF1 cells show that, in the absence of CHX, the S and M footprints from immunopurified ribosomes are markedly underrepresented in approximately the first half of the coding region and become overrepresented in the second half of the coding region compared to total ribosomes (Fig. 4A; Supplemental Figs. S7A, S8A,D). The same pattern holds for the M footprints in libraries from CHX-treated cells (Fig. 4B,C, top panels; Supplemental Figs. S7A, S8B,C, top panels; Supplemental Fig. S8E,F). These observations suggest that Upf1 is not stripped off the coding region by translating ribosomes (Hogg and Goff 2010; Kurosaki and Maquat 2013; Zund et al. 2013) and that Upf1 association with ribosomes occurs routinely during the course of translation elongation. The metagene distribution of S and M footprints recovered from immunopurified ribosomes was nearly identical regardless of the presence or absence of CHX (Fig. 4D, bottom panel), except for a small peak over the start codon in M footprints in either condition (Supplemental Fig. S7A), indicating that the observed progressive increase in Upf1-associated ribosomes that form S and M footprints across the coding region is not CHX-dependent.
FIGURE 4.
Upf1 progressively associates with ribosomes across mRNA coding regions except when Upf1 association forms L footprints. Distribution of footprint abundance across the coding region (CDS) for each footprint size class from total (purple) or IP (orange) ribosomes profiling libraries. (A) Small (S) and medium (M) footprint distribution from strains expressing WT + EV and FLAG-UPF1 in the absence of CHX treatment. (B) Medium (M) and large (L) footprint distribution from strains expressing WT + EV and FLAG-UPF1 in the presence of CHX. (C) Medium (M) and large (L) footprint distribution from CHX-treated cells expressing WT + EV, UPF1-FLAG, UPF1-FLAG/upf2Δ, or upf1DE572AA-FLAG. Each gene's CDS was divided into 100 bins and the percentage of footprints’ P-sites belonging to each bin in all genes was calculated. Gray dashed line signifies a theoretical number where each percentage of CDS contains an equivalent number of footprints for a sum of 100% across all 100 bins. (D) Distribution of dominant footprint (red = Medium, M for +CHX; green = Small, S for −CHX) abundance across the coding region. Data is the same as used in A and B top panels but replotted in such a way to directly observe the differences or similarity in footprint distribution between the absence and presence of CHX during the library preparation of each strain.
The L footprints, which are only detectable with Upf1-associated ribosomes in CHX-treated cells, accumulate rapidly at the beginning of mRNA coding regions and slowly decrease their relative accumulation across the entire coding region in libraries prepared from immunopurified ribosomes from FLAG-UPF1, UPF1-FLAG, or UPF1-FLAG/upf2Δ strains (Fig. 4B,C, bottom panels; Supplemental Fig. S8B,C, bottom panels; Supplemental Table S4). The relative accumulation of L footprints toward the 5′ end of mRNAs is significantly higher than the relative accumulation of M footprints (Supplemental Table S4). These patterns, and their expected 3-nt periodicity, are also evident in higher resolution metagene plots of the first and last 100 nt of the coding regions (Supplemental Fig. S7). Notably, L footprints do not accumulate over the start codon as do M footprints (Supplemental Fig. S7, green lines). The rapid appearance of L footprints relative to ribosome progression across mRNA coding regions (Fig. 4; Supplemental Fig. S7) indicates that their formation commences early during translation and is either stabilized by the CHX-mediated elongation block or that Upf1 may prefer to bind to ribosomes which are in the pretranslocation state trapped by CHX (Wu et al. 2019; Zhao et al. 2021).
Analysis of mRNAs enriched in Upf1-associated ribosomes supports Upf1's role in mRNA surveillance
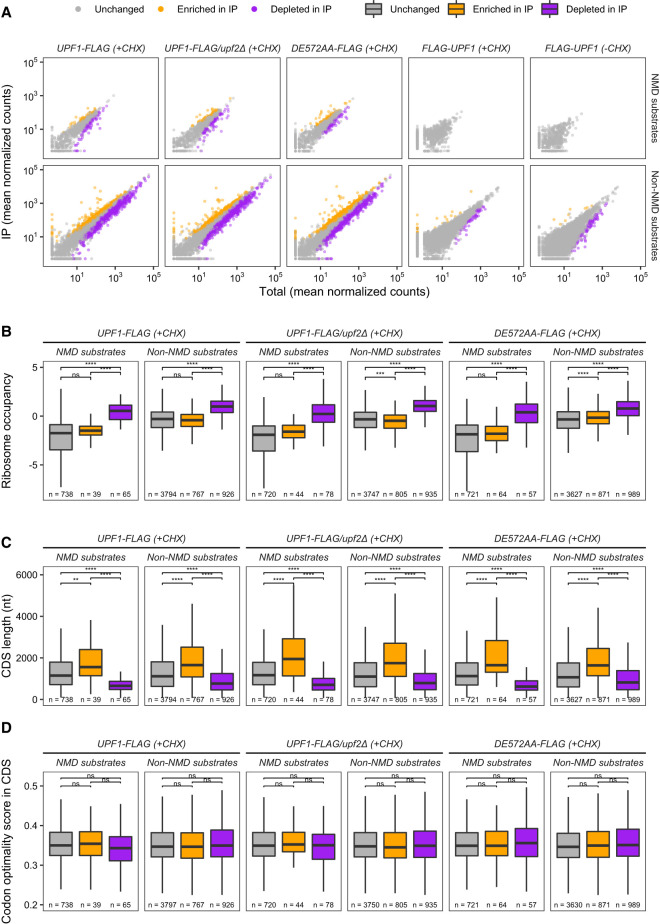
To gain insight into the targets of Upf1 binding, we used DESeq2 to compare the distribution of reads in libraries prepared from IP'd and total ribosomes and identified mRNAs enriched, depleted, or unchanged in IP libraries. We found a larger number of mRNAs identified as having significant differences between IP'd and total ribosomes from the carboxy-terminally FLAG-tagged strains than those from amino-terminally FLAG-tagged strains (Fig. 5A). Because the false discovery rate (FDR) in differential expression analyses takes into account replicate variability, the amino-terminally FLAG-tagged data set, which had two instead of three biological replicates and smaller sequencing library size than the carboxy-terminally FLAG-tagged data set (Supplemental Table S1), had higher variability between replicates, resulting in a smaller number of mRNAs with significant adjusted P-values compared to the carboxy-terminally FLAG-tagged data set. Thus, we focused on the carboxy-terminally FLAG-tagged data set for subsequent analyses.
FIGURE 5.
Characteristics of mRNAs enriched or depleted for Upf1-associated ribosomes. (A) Results of differential expression analyses by DESeq2 between mRNA abundance in IP versus Total ribosome profiling libraries of each strain and CHX treatment. False discovery rate (FDR) with a threshold of 0.05 was used to determine significant differential expression. mRNAs with adjusted P-value <0.05 and positive log2(IP/Total) were considered enriched in IP libraries (orange), whereas those with adjusted P-value <0.05 and negative log2(IP/Total) were considered depleted in IP libraries (purple); otherwise, their abundance did not differ (“unchanged”) between IP and Total (gray). (B–D) Comparative analyses of ribosome occupancy (B), coding sequence length (C), and codon optimality score (D) between mRNA groups identified in A. Two-tailed Wilcoxon's rank sum test with FDR method for multiple testing correction was used to compare each pair of mRNA groups. Symbols for levels of significance are ns: P > 0.05, (*) P < 0.05, (**) P < 0.01, (***) P < 0.001, (****) P < 0.0001. Number of mRNAs in each group is provided. Outlier mRNAs (those beyond the whiskers of box plots) were included in the analyses but omitted from plotting.
In the carboxy-terminally FLAG-tagged data set, we found that both NMD substrates and non-NMD substrates can be enriched, depleted, or unchanged in all IP'd samples (Fig. 5A). These observations suggest that Upf1 can bind ribosomes translating any mRNA and that Upf1 binding to a ribosome by itself does not trigger NMD of the associated mRNA. However, the enrichment and depletion of some transcripts after IP suggests that specific mRNA features may enhance or reduce recruitment of Upf1 to ribosomes.
To characterize mRNAs enriched or depleted in the libraries from immunopurified ribosomes, we performed comparative analyses of ribosome occupancy, coding sequence length, and codon optimality between mRNAs enriched, depleted, or unchanged in IP versus total ribosomes from UPF1-FLAG, UPF1-FLAG/upf2Δ, and upf1DE572AA-FLAG strains (Fig. 5B–D). Consistent with our earlier observations that NMD substrates have significantly lower ribosome occupancy than non-NMD substrates (Celik et al. 2017), we found that mRNAs enriched in Upf1-associated ribosomes for both NMD substrate and non-NMD substrate categories in all strains have significantly lower ribosome occupancy than those depleted in Upf1-associated ribosomes (Fig. 5B; Celik et al. 2017; He et al. 2018). This observation suggests that Upf1:ribosome association occurs more frequently when an mRNA's translation dynamics are similar to those of NMD substrates, and less frequently when an mRNA's translation dynamics are opposite those of NMD substrates. However, the fact that this pattern is observed in both NMD and non-NMD substrates and in strains in which the NMD pathway is inactivated (UPF1-FLAG/upf2Δ and upf1DE572AA-FLAG) further suggests that Upf1 binding to ribosomes is likely to be a surveillance step prior to NMD commitment.
NMD activation is triggered by premature termination events, including out of frame translation ending at a premature stop codon, an event that may occur by chance more frequently in mRNAs with relatively long open reading frames. Therefore, we analyzed the coding sequence length for all mRNAs recovered in our ribosome profiling libraries. We found that mRNAs enriched in IP have significantly longer coding sequence lengths than mRNAs in both the unchanged and depleted groups and that mRNAs depleted in IP have significantly shorter coding sequence lengths than mRNAs in the other two groups (Fig. 5C). However, there was no difference in these patterns between NMD and non-NMD substrates nor between strains with an active or inactive NMD pathway. As such, it is unlikely that this observation is related to Upf1's NMD function but rather reflects the steady ribosomal accumulation of Upf1 across the coding region observed in Figure 4; Supplemental Figures S7, S8, and the increased opportunities for Upf1 association on longer coding sequences, a result that again supports Upf1:ribosome association prior to NMD commitment.
Previous studies (Presnyak et al. 2015; Radhakrishnan et al. 2016) showed that mRNAs targeted for decay tend to have a high number of nonoptimal codons, with corresponding decoding tRNAs that are low in abundance leading to slow elongation. We also showed previously that NMD substrates have slightly lower codon optimality scores than non-NMD substrates (Celik et al. 2017). Thus, we wondered whether this trend is also true for transcripts enriched in IP libraries. We calculated a codon optimality score for each transcript by determining the geometric mean of codon optimality scores of individual codons in the coding sequence (dos Reis et al. 2004; Tuller et al. 2010b). We found no difference in mean codon optimality scores among the three mRNA groups in any strains (Fig. 5D), again supporting our hypothesis that Upf1 associates generally with ribosomes while playing a surveillance function prior to NMD commitment. It is possible, however, that Upf1 could be recruited to localized regions of nonoptimal codons in an mRNA, and thus a single codon optimality score of the entire coding region cannot capture this phenomenon.
Ribosomes associated with upf1DE572AA accumulate downstream from normal termination codons
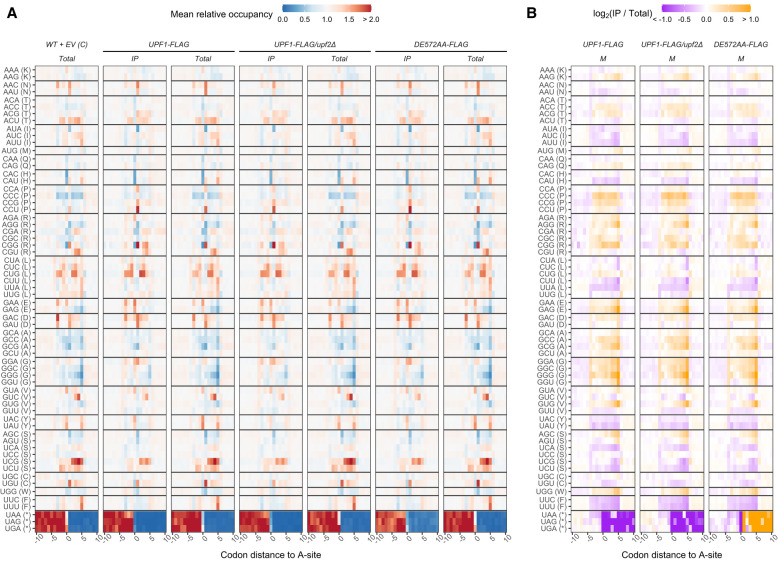
To determine whether Upf1-associated ribosomes are found at specific codons, we calculated the mean relative A-site codon occupancy of immunopurified and total ribosomes for any given codon and its surrounding region (Fig. 6A). Because ribosomal pauses are related to translation elongation dynamics and the abundance of tRNAs in the cell (Tuller et al. 2010a; Dana and Tuller 2014), and their relationships have been shown to be disrupted by CHX treatment (Hussmann et al. 2015; Sharma et al. 2021), we tested whether CHX affects codon occupancy analyses in our libraries by computing Spearman's rank correlation of A-site mean relative occupancy with the inverse of tRNA adaptation index (tAI) (dos Reis et al. 2004; Tuller et al. 2010b) for a given sense codon identity in each library, as described by Hussmann et al. (2015) (Supplemental Fig. S9). Consistent with results from the Hussmann et al. (2015) analysis, we found positive correlations for both S and M footprints for libraries untreated with CHX (Supplemental Fig. S9, open circles), demonstrating that the expected relationship between translation dynamics and codon optimality is maintained. Moreover, we found negative correlations for both M and L footprints for libraries derived from cells treated with CHX (Supplemental Fig. S9, circles with plus sign), indicating that CHX disrupted the ability to accurately measure the impact of codon optimality on ribosomal enrichment or depletion at sense codons.
FIGURE 6.
Codon occupancy for individual codon identity in the A-site and its surrounding codon positions in carboxy-terminally FLAG-tagged UPF1 libraries. (A) Mean relative occupancy values were calculated as described in Materials and Methods. Briefly, read count of major footprint size (M in this case) at each codon position relative to the codon of interest in the A-site was normalized by the average count in the 60-codon window. Thus, mean relative occupancy of 1 (white) indicates that A-site occupancy at that position is no different from the window average. Mean relative occupancy >1 (red) and <1 (blue), respectively, indicate enrichment and depletion of ribosomal A-site at that position compared to window average. Only codons −10 to 10 were plotted. (B) Differences in mean relative occupancy between IP and total libraries. Ratios of mean relative occupancy (from A) of IP to Total, log2-transformed. Thus, log2(IP/Total) of 0 (white) indicates no difference in mean relative occupancy between IP and Total at that codon position, while positive (orange) and negative (purple) log2(IP/Total) indicate, respectively, higher and lower occupancy in IP compared to total at that codon position.
When we compared log2 fold change in mean relative occupancy of immunopurified versus total ribosomes across libraries prepared from the three carboxy-terminally FLAG-tagged strains, we found that their patterns of enrichment or depletion around stop codons vary between strains (Fig. 6B). In strains expressing WT UPF1-FLAG, whether in UPF2 or upf2Δ background, we found that mean relative occupancy of immunopurified ribosomes were depleted compared to total ribosomes in the 3′-UTR region downstream from all three in-frame stop codons (Fig. 6B, purple highlighting). In contrast, we found that in libraries from strains expressing upf1DE572AA-FLAG, the mean relative occupancy of immunopurified ribosomes in the 3′-UTR region was increased compared to total ribosomes (Fig. 6B, orange highlighting). These differences in the amount of 3′-UTR footprints between IP and total libraries were also evident when we calculated the overall percentage of footprints in the libraries that mapped to the 3′-UTR region (Fig. 7A; Supplemental Fig. S10B). Unlike other strains containing WT UPF1, where the 3′-UTR footprints were slightly reduced in IP compared to total libraries, the upf1DE527AA-FLAG strain IP libraries show an increase to almost double the amount of 3′-UTR footprints compared to total libraries, and this increase is statistically significant (Fig. 7A; Supplemental Fig. S10B). The upf1DE572AA mutation has been observed to produce mRNP-protected decay intermediates downstream from premature stop codons in transcripts from reporter constructs (Franks et al. 2010; Serdar et al. 2016; Serdar et al. 2020), and these decay intermediates were later found to be bound by a ribosome at their 5′ terminus (Serdar et al. 2020). Our observation that footprints from Upf1-bound ribosomes are selectively enriched downstream from normal termination codons in the upf1DE572AA-FLAG strain is surprising in light of the generally accepted notion that Upf1 functions during premature termination (He and Jacobson 2015; Kurosaki et al. 2019).
FIGURE 7.
Percentage of footprints in the 3′-UTR region. (A) Percentage of footprints whose P-sites fall into the 3′-UTR region for each library. (B) Percentage of frame 0 footprints within the 3′-UTR region. Horizontal gray dashed line indicates a theoretical percentage of 33% where all three reading frames would be equally represented. For both A and B, percentage calculated from each replicate was plotted as a gray dot. Bar plot represents the average among the three replicates. Two-tailed paired t-test was used to determine significant differences between percentages from IP and Total libraries.
Footprints in the 3′-UTR of an mRNA can be caused by several events, such as nonsense codon readthrough, reinitiation, and frameshifting. Previously, ribosomes associated with 3′ decay intermediates in the upf1DE572AA mutant strain were determined not to be engaged in canonical translation, neither readthrough nor reinitiation (Serdar et al. 2020). Therefore, their footprints would be expected to have a random reading frame and the percentage of in-frame 3′-UTR footprints in IP libraries prepared from the upf1DE572AA-FLAG strain should be reduced compared to total ribosome libraries or libraries prepared from strains harboring WT UPF1. Thus, we calculated percentages of the three reading frames in the 3′-UTR region of all libraries (Fig. 7B; Supplemental Fig. S10D) and used values from the total ribosome libraries to establish baseline level of these events. We found that from total libraries of all carboxy-terminally FLAG-tagged strains, reading frame 0 accounts for ∼50%–55% of the footprints in the 3′-UTR. In IP libraries of strains containing UPF1-FLAG, the percentage of frame 0 footprints is reduced slightly to ∼45%–49%. However, in IP libraries of upf1DE572AA-FLAG strain, the percentage of frame 0 drops to 37%, approximately equivalent to frames 1 and 2. This evidence suggests that these footprints, like the 3′ decay intermediates observed previously (Serdar et al. 2020), are not generated by ribosomes undergoing canonical translation but rather are a consequence of downstream ribosome migration and defective ribosome recycling attributable to ribosome-bound ATPase-deficient Upf1. Further, while we recognize that this unexpected detection of Upf1's action at normal termination codons could be due to episomal expression of UPF1, the detection of these footprints only in IP libraries of upf1DE572AA-FLAG cells supports the biological significance of our findings.
DISCUSSION
Upf1 functions while associated with ribosomes
Given the uncertainties underlying the association of Upf1 with elongating ribosomes (see Introduction), we sought to elucidate the issue by combining UPF1 genetics with selective ribosome profiling. While recognizing the caveat that our observations may be influenced by episomal expression of Upf1, the results of these experiments strongly support the notion that Upf1 functions while associated with ribosomes engaged in elongation. In support of this conclusion we have shown that: (i) immunopurification of FLAG-tagged Upf1 from a sample of total cellular ribosomes yields stoichiometric recovery of Upf1 and ribosomal proteins that is specific for the FLAG tag regardless of its 5′ or 3′ location in the respective UPF1 ORF or the status of NMD activity in the cells from which ribosomes were isolated (Fig. 1); (ii) Upf1-associated ribosomes purified from cells treated or not treated with CHX, respectively, yield the expected predominant 27–32 nt (“M”) or 20–23 nt (“S”) footprint size classes and these footprints manifest appropriate 3 nt periodicity characteristic of translating ribosomes (Fig. 2; Supplemental Fig. S7); (iii) Upf1 association with ribosomes in CHX-treated cells leads to formation of an additional 5′-extended 37–43 nt (“L”) footprint and it, too, manifests 3 nt periodicity (Figs. 2, 3; Supplemental Fig. S7; see also next section); (iv) S and M footprints from immunopurified ribosomes are relatively underrepresented in approximately the first half of normalized coding regions and overrepresented in the second half (Fig. 4; Supplemental Fig. S8) whereas L footprints rapidly accumulate in normalized ORFs and maintain their presence throughout mRNA coding regions, diminishing only slightly from the start of translation until its termination (Fig. 4; Supplemental Fig. S8); and (v) ribosomes associated with Upf1 that harbors the DE572AA amino acid substitutions terminate or recycle improperly at normal termination codons and are retained in mRNA 3′-UTR regions (Figs. 6, 7; Supplemental Fig. S10). These results lead us to conclude that, at least in yeast, Upf1 is not displaced by elongating ribosomes, but is carried by them routinely during the course of translation elongation. Since NMD substrates and non-NMD substrates were found to be enriched, depleted, or unchanged in all immunopurified ribosome samples (Fig. 5) it appears that stochastic binding of Upf1 to translating ribosomes is insufficient to trigger NMD of the associated mRNA and that the observed association may well be part of a translation surveillance mechanism in which Upf1 is sensing termination efficiency.
The Upf1:ribosome complex detected in CHX-treated cells comprises a step that precedes commitment to NMD
In CHX-treated cells, the footprints recovered from immunopurified Upf1-associated ribosomes include the expected (Wu et al. 2019) M footprints of ∼27–32 nt as well as novel 37–43 nt ribosome-protected fragments that we designate as L footprints (Fig. 2). The latter footprints may be formed by a CHX-induced ribosome collision and subsequent endonucleolytic cleavage (Guydosh and Green 2017) or by the combined mRNA protection effects of Upf1 plus a ribosome to which it is bound (Fig. 3). The latter additive effect would be consistent with the size of ribosome footprints and the observation that human Upf1 protects 8–11 nt of RNA in RNA binding and unwinding assays (Chakrabarti et al. 2011). L footprints accumulate early during translation of mRNA ORFs, well before distal regions of mRNA are recovered in M or S footprints from Upf1-associated ribosomes (Fig. 4; Supplemental Fig. S8). L footprints are not recovered from Upf1-associated ribosomes purified from cells without CHX treatment (Fig. 2A), and their formation does not depend on specific placement of the FLAG epitope tag to one end of Upf1 (Fig. 2). Formation of L footprints does depend on Upf1 function because they are absent when Upf1 harbors DE572AA substitutions, but they are not dependent on a functional NMD pathway or Upf2 activity because they are still recovered from Upf1-associated ribosomes in CHX-treated upf2Δ cells (Fig. 2B). This combination of properties suggests that CHX treatment has trapped ribosomes and Upf1 in an otherwise transient state preceding a commitment to NMD, that is, L footprints are Upf1-specific, not NMD-specific. The existence of such a transient state implies that Upf1 can interact with ribosomes in modes that either do or do not lead to NMD. The early appearance of L footprints (i.e., their recovery from 5′ regions of mRNA ORFs), and their persistence throughout most of the coding region (Fig. 4; Supplemental Fig. S7), implies that this potential surveillance for functional targets (e.g., a ribosome undergoing premature termination) is likely to be active during most translation elongation events. In addition, the uniformly delayed recovery of S and M footprints (the majority of the footprints recovered) from Upf1-associated ribosomes (Fig. 4; Supplemental Fig. S7), is synchronous with the onset of decline in L footprint formation, suggesting distinct states of Upf1 interaction with ribosomes.
ATPase-deficient Upf1 interferes with normal translation termination
Our analyses of the mean relative codon occupancy of immunopurified vs. total ribosomes in libraries from the carboxy-terminally FLAG-tagged strains (treated with CHX) indicated that the relative ribosomal enrichment or depletion around all three stop codons was not comparable for the three strains (Fig. 6B). Libraries prepared from immunopurified ribosomes derived from cells expressing WT UPF1-FLAG, with or without functional Upf2, were quite similar and manifested a relative depletion of ribosomes in the 3′-UTR region downstream from all three in-frame stop codons (Fig. 6B, purple highlighting). This result was consistent with ongoing translation termination and ribosome release at the normal ends of essentially all mRNA ORFs. In contrast, cells expressing upf1DE572AA-FLAG manifested a deficiency in normal termination and ribosome release, that is, the accumulation of ribosomes downstream from normal termination codons without regard to reading frame (Figs. 6B, 7A; Supplemental Fig. S10B).
Upf1 was originally identified as a regulator of the stability of mRNAs undergoing premature translation termination (Leeds et al. 1991; He et al. 1993) and this role for Upf1 has been substantiated by three decades of additional studies (He and Jacobson 2015; Kurosaki et al. 2019) that describe a cascade of Upf1-driven events commencing with premature termination and culminating with mRNA decapping or endonucleolytic cleavage and subsequent exonuclease digestion (Muhlrad and Parker 1994; He and Jacobson 2001, 2015; Huntzinger et al. 2008; Eberle et al. 2009; Loh et al. 2013; Colombo et al. 2017; Ottens et al. 2017; Nelson et al. 2018; Kurosaki et al. 2019; He et al. 2022). It was thus surprising that cells expressing upf1DE572AA exhibited an apparent defect in normal termination, particularly since a recent study of reporter mRNAs containing premature termination codons (PTCs) in upf1DE572AA yeast cells also accumulated unreleased ribosomes downstream from stop codons, but these stop codons were all PTCs (Serdar et al. 2020). Our results are consistent with the notion that ribosome-bound Upf1 normally monitors the status of translation termination events and exerts its activity only when those events are aberrant, that is, when they occur prematurely or in premature context, but suggest that the DE572AA mutation may in some instances interfere with normal termination as well as premature termination. For example, the Upf1 DE572AA mutant protein may be able to target elongating ribosomes but cannot dissociate from them efficiently because of its disrupted ATPase cycle. This mutant Upf1 protein could thus persist on ribosomes, even at normal termination codons, and interfere with normal ribosome recycling, causing ribosome accumulation in mRNA 3′-UTRs.
Upf1 functions in multiple complexes
Finally, there remains the question of the specific function of ribosome-bound Upf1 in the NMD pathway. Our earlier results suggested that Upf1 may promote the release of an otherwise poorly dissociable premature termination complex (Ghosh et al. 2010). While the results of Figures 6, 7, and Supplemental Figure S10 support this possibility, our recent study on the role of decapping activators in the targeting and activation of the decapping enzyme suggests an additional function (He et al. 2022). In that study, we showed that specific cis-binding elements in the Dcp2 carboxy-terminal domain control the substrate specificity of the decapping enzyme by orchestrating the formation of target-specific decapping complexes. The latter include a Upf1-containing decapping complex that targets NMD substrates (He et al. 2022). Since deletion of UPF1 promotes extensive stabilization of NMD substrates but Upf1-mediated recruitment of the decapping enzyme only makes a minor contribution to the overall decay of NMD substrates (He et al. 2022), it is likely that Upf1 carries out a major function upstream of decapping enzyme recruitment. In light of the fact that Upf1 has two nearly identical binding sites in the unstructured Dcp2 carboxy-terminal domain (He et al. 2022), and that Upf1 is known to dimerize (He et al. 2013), it is possible that ribosome-associated Upf1 remodels an mRNP to render it susceptible to decapping and then facilitates decapping by dimerizing with a monomer of Upf1 present in an NMD-specific decapping complex. Consistent with this proposition, Upf1 has been found in two distinct complexes, a “detector” complex containing Upf1, Upf2, and Upf3, and an “effector” complex containing Dcp1, Dcp2, and Edc3, as well as Nmd4 and Ebs1 (Dehecq et al. 2018).
MATERIALS AND METHODS
Yeast strains
Yeast strains used in this study are listed in Supplemental Table S5. Strains containing complete gene deletions of UPF1 and UPF2 (HFY871, HFY861, HFY467) were described previously (He and Jacobson 1995; He et al. 1997).
Oligonucleotides
Oligonucleotides used in this study were obtained from Eurofins Operon or Integrated DNA Technologies (IDT) and are listed in Supplemental Table S6. Gene fragments were synthesized by Quintara Biosciences and are described below in Plasmids.
Plasmids
Plasmids used in this study are listed in Supplemental Table S7. The following plasmids were published previously: YEplac112 (Gietz and Sugino 1988); pG1-FLAG-UPF1 (Czaplinski et al. 1995); pRS425 and pRS316(Sikorski and Hieter 1989).
YEplac112-6Xhis-UPF1: pRS314-UPF1 (He and Jacobson 1995) was digested with BamHI, SalI, and XbaI to release a 4.2kb BamHI-SalI fragment of UPF1.
This fragment was ligated into pGEM-3Zf (+) (Promega) that had been digested with BamHI and SalI and dephosphorylated with calf intestinal alkaline phosphatase (NEB). 6Xhis tag was added to the amino terminus of Upf1 in pGEM-3Zf (+) using a QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent) with oligonucleotides 5-N-His-UPF1 and 5-N-HIS-UPF1-r to yield pGEM3Zf(+)-6Xhis-UPF1. A ∼4.2 kb SacI/PstI fragment was isolated from pGEM3Zf(+)-6Xhis-UPF1, ligated into YEplac112 which had been digested with SacI and PstI and dephosphorylated with calf intestinal alkaline phosphatase to yield YEplac112-6Xhis-UPF1.
pRS425-UPF1-FLAG: gene fragments were synthesized by Quintarabio with flanking restriction sites (i) UPF1-Xho-Sph, a 1593 bp fragment containing the TDH3 (glyceraldehyde-3-phosphate dehydrogenase, GAPDH) promoter inserted into a BamHI site plus ggc (ggatccggc) immediately upstream of the start codon of the UPF1 coding region, then from the start codon to the SphI site at position 914 of the UPF1 coding sequence; (ii) UPF1-Sph-Nco, a 1102 bp fragment from the SphI site in the UPF1 coding sequence to the NcoI site in the UPF1 coding sequence; (iii) UPF1-Nco-Sac, a 1167 bp fragment from the NcoI site in the UPF1 coding sequence to a SacI site in the UPF1 3′UTR followed by a spacer with a SacI restriction site (caccgcggtggagctc), and FLAG epitope sequence (gattacaaggatgacgacgataag) inserted immediately upstream of the stop codon. These three gene fragments were ligated into pRS425 at the XhoI-SacI sites of the MCS.
Mutations of UPF1 were placed into UPF1-FLAG as follows:
C62Y: The UPF1-Sph-Nco and UPF1-Nco-Sac gene fragments described above were cloned in to the SphI-SacI sites in the pGEMT-EZ (Promega) MCS to generate pGEMT-EZ-Sph-UPF1-FLAG-Sac.The UPF1-Xho-Sph gene fragment in plasmid vector pQ (Quintarabio) was mutagenized with oligonucleotides 5Upf1C62Ymut and 3Upf1C62Ymut using the QuikChange II XL Site-directed mutagenesis kit (Agilent). The UPF1XhoI-C62Y-SphI fragment was isolated and ligated with the 2.28 kb Sph-UPF1-FLAG-Sac fragment from pGEMT-EZ-Sph-UPF1-FLAG-Sac into the XhoI-SacI site of the MCS of pRS425.
K436E: pGEM3Z-6Xhis-UPF1 was mutagenized with oligonucleotides K436F and K436R using the QuikChange II XL Site-directed mutagenesis kit; the 1116 bp SphI-NcoI fragment containing the K436E mutation was isolated and substituted for the UPF1-Sph-Nco fragment as described above for the construction of pRS425-UPF1-FLAG.
AKS484HPA, RR793AA and R779C: pGEMT-EZ-Sph-UPF1-FLAG-Sac was mutagenized with oligonucleotides Upf1AKS-HPAmut and UPF1AKS-HPAmutrev, 5Upf1RRAAmut and 3Upf1RRAAmut, or Upf1R779C and Upf1R779Crev, respectively, using the QuikChange II XL Site-directed mutagenesis kit. The 2.28 kb SphI-SacI fragments were isolated and were ligated with the UPF1-Xho-Sph gene fragment into the XhoI-SacI site of the pRS425 MCS.
DE572AA: the 1116bp SphI-NcoI fragment from pACTII-UPF1-TH4-3-2-DE572AA (He et al. 2013) was isolated and substituted for the UPF1-Sph-Nco fragment described above.
UPF1-FLAG and mutants were placed under the UPF1 promoter into pRS316 as follows: a 340 bp fragment containing the UPF1 promoter sequence flanked by XhoI and BamHI restriction sites was generated by PCR from pRS313-UPF1 (E-B) with oligonucleotides 5Upf1XhoProm and 3Upf1BamProm and digested with XhoI and BamHI. A 3.195 kb BamHI-SacI fragment from pRS425-UPF1-FLAG or mutants was isolated and ligated with the digested PCR product above into the XhoI-SacI site of the pRS316 MCS.
pRS425-UPF2: a 4946 bp fragment generated by PCR with primers Upf2-425-fwd and Upf2-425-rev containing a the entire UPF2 gene was inserted into pRS425 cut with NotI and SalI using the NEBuilder High-Fidelity DNA Assembly Cloning Kit (New England Biolabs).
YEplac195-TPI-UPF3: A 1164 bp SalI-XbaI fragment was generated by PCR from genomic DNA with primers 5′Sal-UPF3 and 3′Xba-UPF3 and restriction digested with SalI and XbaI. YEplac195-TPI-FLAG-UPF3 was digested with SalI and XbaI and the vector backbone was ligated to the digested PCR product to generate YEplac195-TPI-UPF3.
Polyribosome analysis, protein detection, and quantitation
For polyribosome analysis, cells were grown in selective media and harvested as described previously (Mangus and Jacobson 1999). Lysates were loaded onto 7%–47% sucrose gradients, subjected to ultracentrifugation, fractions collected, and individual fractions were precipitated with trichloroacetic acid (TCA). Samples from total or immunopurified ribosomes or aliquots from TCA precipitated sucrose gradient fractions (fractions 1–10, undiluted; fractions 11 and 12, diluted 1:10) were run in 1× Laemmli buffer on 4%–20% Mini-PROTEAN TGX Precast Protein Gels (Bio-Rad) in 1× Tris/Glycine/SDS buffer (Bio-Rad), subjected to western blotting onto Immobilon-P PVDF Transfer Membranes (EMD Millipore) by electrotransfer using a Trans-Blot SD Semi-Dry Transfer Cell (Bio-Rad). Protein was visualized with rabbit α-FLAG polyclonal antibody (Sigma) or rabbit monoclonal α-Rps6 antibody (Cell Signaling) followed by HRP-conjugated donkey α-rabbit (GE) antibody. Polysomal distribution of tagged WT and mutant Upf1 protein and tagged protein signal from total or immunopurified ribosomes was visualized by western blotting on Amersham Hyperfilm ECL film (GE). Band quantitation from scanned images was performed using Multigauge V3.0 (Fuji). For quantitative analysis of the polysome distribution of tagged proteins, the antibody signal for a given fraction was calculated as the percent of its total signal across fractions 1–10 of an individual sucrose gradient western blot. Values from western blots of replicate gradient fractions were averaged and a bar graph generated to display the mean percent signal across sucrose gradient fractions 1–10 from all replicates of that sample with standard error of the mean displayed.
Ribosome purification
Yeast cultures were grown, whole-cell yeast lysates were prepared and digested with RNaseI, and ribosomes were recovered as described previously (Ganesan et al. 2019).
Negative-stain transmission electron microscopy
Ribosomes were prepared for electron microscopy using the conventional negative staining procedure. Formvar-carbon-coated copper grids were treated by glow discharge using a PELCO easiGlow glow discharge cleaning system for 30 sec at 20 mA. The ribosomes were diluted in footprinting buffer (20 mM Tris, pH 7.4, 150 mM NaCl, 5 mM MgCl2) and negatively stained with 1% (w/v) aqueous uranyl acetate. All images were taken with an FEI Tecnai Spirit 12 (FEI Company), operated at 120 kV. Micrographs were recorded with a Gatan Rio 9 CMOS camera.
Immunopurification
FLAG-tagged protein-associated ribosomes were isolated by immunopurification using anti-FLAG M2 Affinity gel as described previously (Ganesan et al. 2019). Eluate was collected by centrifugation and spun through Amicon Ultra 2 mL 100K centrifugal filter units (Millipore) until the volume was <200 µL. The concentrate from one reaction set was pooled into one Amicon Ultra 2 mL 100K centrifugal filter unit and spun until the total volume was ∼160 µL. For the UPF1-FLAG, upf1DE572AA-FLAG and UPF1-FLAG/upf2Δ ribosome profiling libraries, immunopurified ribosomes were flash frozen and stored at −80°C until RNA purification. For FLAG-UPF1 libraries and for all mass spectrometry analyses, the immunopurified concentrate was layered on top of a 1 M sucrose cushion in IP buffer plus 1× protease inhibitors, 10 U/mL Superase-In, and spun at 166,180g for 100 min at 4°C in a TLA100 rotor. Supernatant was removed and the pellet was resuspended in 60 µL IP buffer plus 0.5 mM DTT, 1× protease inhibitors, 20 U/mL Superase-In; and A260 was determined on a spectrophotometer.
Sample preparation for mass spectrometry
Mass spectrometry analyses were performed by the Mass Spectrometry Facility of the UMass Chan Medical School. Samples were prepared for mass spectrometry by disrupting total or immunopurified ribosomes in 1× Laemmli buffer and running on a 4%–20% Mini-PROTEAN TGX Precast Protein Gel in 1× Tris/Glycine/SDS buffer (Bio-Rad) for 5 min. Gels were stained using the NOVEX Colloidal Blue Staining Kit (Thermo Fisher Scientific) and destained in water. Fragments were excised and cut into 1 × 1 mm pieces, transferred to microfuge tubes, and each added 1 mL of water, followed by a solution of 20 µL of 45 mM of 1,4 dithiothreitol (DTT) in 200 µL of 250 mM ammonium bicarbonate. Samples were incubated at 50°C for 30 min, cooled to room temperature, added 20 µL of 100 mM iodoacetamide (IAA) and incubated for 30 min. Excessive DTT and IAA was removed, and the gel pieces washed with water (3×, 1 mL each), followed by 1 mL of a 1:1 solution of 50 mM ammonium bicarbonate:acetonitrile, quenched with 200 µL of acetonitrile and dried in a Speed Vac. Gel pieces were rehydrated in a mixture of 4 ng/µL trypsin (Promega) and 0.01% ProteaseMAX (Promega) in 50 µL of 50 mM ammonium bicarbonate and incubated for 18 h at 37°C. Supernatants were collected and further extraction was performed by adding 200 µL of 80:20 solution of acetonitrile:1% (v/v) formic acid in water. Supernatants were combined, peptides were lyophilized in a Speed Vac and resuspended in 25 µL of 5% acetonitrile with 0.1% (v/v) formic acid for mass spectrometry analysis.
Mass spectrometry
Data was acquired using a NanoAcquity UPLC (Waters Corporation) coupled to a Q Exactive hybrid mass spectrometer (Thermo Fisher Scientific) (amino-terminal FLAG-tagged data sets) or an Orbitrap Fusion Lumos Tribrid (ThermoFisher Scientific) mass spectrometer (carboxy-terminal FLAG-tagged data sets). Peptides were trapped and separated using an in-house 100 µm I.D. fused-silica precolumn (Kasil frit) packed with 2 cm ProntoSil (Bischoff Chromatography, DE) C18AQ (200 Å, 5 µm) media and configured to an in-house packed 75 µm I.D. fused-silica analytical column (gravity-pulled tip) packed with 25 cm Magic (Bruker) C18AQ (100 Å, 3µm) media, respectively. Mobile phase A was 0.1% (v/v) formic acid in water and mobile phase B was 0.1% (v/v) formic acid in acetonitrile. Following a 3.8 µL sample injection, peptides were trapped at a flow rate of 4 µL/min with 5% B for 4 min, followed by gradient elution at a flow rate of 300 nL/min from 5%–35% B in 60 min, wash and reconditioning (total run time ∼90 min). Electrospray voltage was delivered by liquid junction electrode (1.4 kV) located between the columns and the transfer capillary to the mass spectrometer was maintained at 275°C. Mass spectra were acquired over m/z 300–1750 Da with a resolution of 60,000 (m/z 200), maximum injection time of 30 msec, and an AGC target of 700,000. Tandem mass spectra were acquired using data-dependent acquisition (2 sec cycle) with an isolation width of 1.2 Da, HCD collision energy of 30%, resolution of 15,000, maximum injection time of 100 msec, and an AGC target of 10,000. Biological triplicates of each strain (input and IP) were analyzed in technical triplicate.
Database searches
Raw data were processed using Proteome Discoverer (Thermo Fisher Scientific, version 2.1.1.21) and searched against Uniprot Yeast (downloaded 07/2021) database using Mascot (Matrix Science, version 2.6.2). Search parameters were as follows: full tryptic specificity with up to two missed cleavages; precursor mass tolerance 10 ppm; fragment mass tolerance 0.05 Da; cysteine carbamidomethylation considered as a fixed modification, while protein amino-terminal acetylation, methionine oxidation, peptide amino-terminal (E,Q) pyroglutamate conversion, serine/threonine phosphorylation and lysine ubiquitination (GG) were specified as variable modifications. Peptide and protein validation and annotation was done in Scaffold 4.8.9 (Proteome Software, Portland, OR) using Peptide Prophet (Keller et al. 2002) and Protein Prophet (Nesvizhskii et al. 2003) algorithms. Peptides were filtered at a 1% FDR, while protein identification threshold was set to greater than 99% probability and with a minimum of two identified peptides per protein. Intensity-based absolute quantification (iBAQ) (Schwanhausser et al. 2011, 2013), calculated and normalized across biosamples in Scaffold, is used as a measure of protein abundance. Protein identification probability, Total Spectrum Count, and normalized iBAQ values for each protein in each sample are provided as Supplemental Tables S8–S11.
Protein abundance analysis
Differential abundance analysis of protein levels between IP and Total ribosomes was performed in R environment using data exported from Scaffold. For proteins with identification probability <99% and Total Spectrum Count <2, their iBAQs were set to zero. For each protein in a sample, at least two replicates were required to have positive iBAQ values for further differential abundance analysis; otherwise, the identification of the protein was deemed inconsistent. iBAQs of biological replicates were averaged and log10-transformed for plotting Figure 1A. Proteins that were exclusively identified in Total or IP samples (i.e., those with missing values in one of them) were plotted on the axes. The list of proteins with their average iBAQs and whether they are exclusively identified in Total or IP or identified in both are available as Supplemental Table S2. For proteins that were identified in both Total and IP samples, their difference in abundance was further analyzed using an R package limma (Smyth 2004; Kammers et al. 2015). First, iBAQs in each replicate samples were log10-transformed. Missing iBAQ values within a sample were empirically imputed from the left-shifted Gaussian distribution of iBAQs of that replicate sample. The model was fit using limma, and the resulting log2 fold change between IP and Total protein abundance with the associated P-value adjusted by the Benjamini–Hochberg method were used to plot Figure 1B. The result tables generated by limma are available as Supplemental Table S3.
mRNA decay analysis
mRNA decay phenotypes were assessed by northern blotting of total RNA prepared from cell pellets by the hot phenol method (Herrick et al. 1990), probing for CYH2 mRNA and pre-mRNA as described previously (Herrick et al. 1990; He et al. 1993; He and Jacobson 1995). Blots were visualized on a Fuji phosphorimager and quantitated with Multigauge V3.0 (Fuji). Northern blot images in Supplemental Figure S3 were subjected to a Multigauge noise reduction filter, kernel size 3 × 3, applied equally across the entire image for display purposes only.
RNA-seq and ribosome profiling library preparation from cells expressing amino-terminal FLAG-tagged Upf1 protein
RNA was isolated from clarified lysates (60 µL), and total (15 µL) or immunopurified ribosomes (entire recovered volume) using the miRNeasy mini kit (Qiagen) according to manufacturer's standard protocol. RNA was eluted in 30 µL (lysate) or 14 µL (ribosomes), then treated with RiboZero Magnetic Gold (Yeast) Kit (Illumina) as described previously (Ganesan et al. 2019). Ribosome protected RNA fragments were 3′ dephosphorylated with T4 polynucleotide kinase (NEB) in 150 mM MES-NaOH pH 5.5, 450 mM NaCl, 15 mM MgCl2, 15 mM β-mercaptoethanol, 0.3 U/µL Superase-In at 37°C, 2 h; 65°C, 20 min; and RNA was selectively recovered with RNA Clean and Concentrator-5 (Zymo Research). RNA fragments were then 5′ phosphorylated with T4 polynucleotide kinase in T4 PNK buffer, 1 mM ATP, 1 U/µL Superase-In at 37°C, 1 h; 60°C, 10 min; and RNA was selectively recovered with RNA Clean and Concentrator-5 (Zymo Research) according to the manufacturer's protocol. Ribosome profiling libraries were prepared using the NEXTflex Small RNA-Seq Kit v3 (Perkin Elmer).
Total mRNA libraries were prepared using the TruSeq Stranded mRNA LT Sample Prep Kit (Illumina) according to manufacturer's protocol.
Over the course of library preparation, the amounts of RNA and final libraries were quantified by Qubit 3.0 Fluorometer with the Qubit RNA HS Assay Kit (Thermo Fisher Scientific) and the Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific), respectively. Assessments of rRNA depletion, RNA quality, and final libraries were done by a Fragment Analyzer capillary electrophoresis system (Advanced Analytical) at the UMass Chan Medical School Molecular Biology Core Labs (RRID: SCR_018263).
RNA-seq and ribosome profiling library preparation from cells expressing carboxy-terminal FLAG-tagged Upf1 protein
Because the RiboZero Magnetic Gold (Yeast) Kit (Illumina) was discontinued midway through this study, we developed an alternative rRNA depletion method based on oligonucleotide blocking of adapter ligation and cDNA synthesis reactions. To maximize the amount of ribosome protected mRNA fragments input into the libraries, we altered immunopurification and RNA isolation protocols for the carboxy-terminally FLAG tagged ribosome profiling libraries as follows by: (i) eliminating the final pelleting through a sucrose cushion following immunopurification, to avoid any loss of immunopurified ribosomes, and (ii) using the small (<200 nt) RNA preparation workflow in the miRNeasy kit according to manufacturer's instructions, to remove as much extraneous RNA (either rRNA or large mRNA fragments) as possible prior to ribosome profiling library preparation.
Following RNA isolation, 10 µL RNA from total or immunopurified ribosomes were 3′ dephosphorylated and 5′ phosphorylated as described above (Ganesan et al. 2019) and selectively recovered with RNA Clean and Concentrator-5 (Zymo Research) according to the manufacturer's instructions, using adjusted RNA Binding Buffer diluted 1:1 with ethanol, into 13 µL water. 8.5 µL RNA was incubated in a thermocycler with a preheated lid with 2 µL oligonucleotides (for RNA from total ribosomes) or 1 µL 1:10 diluted oligonucleotides (for RNA from immunopurified ribosomes) from the QIAseq FastSelect –rRNA Yeast Kit (Qiagen) in 10.5 µL total at 75°C, 2 min; 70°C, 2 min; 65°C, 2 min; 60°C, 2 min; 55°C, 2 min; 37°C, 2 min; 25°C, 2 min; 4°C, hold. Ribosome profiling libraries were prepared using the NEXTflex Small RNA-Seq Kit v3 (Perkin Elmer) but eliminating the 70°C denaturation step prior to 3′ 4N Adenylated Adapter ligation. Adapters were undiluted for RNA from total ribosomes and 1:4 diluted for RNA from immunopurified ribosomes. PCR cycles were performed until a library appeared by Fragment Analyzer analysis, 15 cycles for libraries from total ribosomes or 15–18 cycles for libraries from immunopurified ribosomes, and libraries were selectively recovered using the gel-free size selection and cleanup protocol according to manufacturer's protocol.
For RNA-seq library preparation, RNAs from clarified lysates (60 µL) were isolated using the miRNeasy mini kit (Qiagen) according to manufacturer's standard protocol. The use of TruSeq Stranded mRNA LT Sample Prep Kit (Illumina) was altered as follows: 1 µg of RNA, 14.5 µL of Fragment, Prime, Finish mix (FPF), 1 µL of oligonucleotides from the QIAseq FastSelect –rRNA Yeast Kit (Qiagen) in a total of 20.5 µL were incubated in a thermocycler with a preheated lid at 94°C, 8 min; 75°C, 2 min; 70°C, 2 min; 65°C, 2 min; 60°C, 2 min; 55°C, 2 min; 37°C, 2 min; 25°C, 2 min; 4°C, hold; followed by first strand cDNA synthesis and the remainder of the protocol according to manufacturer's instructions, except RNA Adapters were diluted 1:2 at the ligation step. If excess adapter dimers were present, it was necessary to reconstruct the library and dilute the RNA Adapter 1:4 or 1:8 and increase the PCR cycle number to 17, as was done for libraries T5019, T5033, T5035, and T5338.
High-throughput sequencing of RNA-seq and ribosome profiling libraries
RNA-seq libraries were sequenced on either a HiSeq4000 (Illumina) at Beijing Genomics Institute (BGI) or a NextSeq500 (Illumina) in-house. Ribosome profiling libraries were sequenced on a NextSeq500 in-house.
Sequence alignment, transcript quantification, and data analyses
RNA-seq and ribosome profiling reads were aligned to a yeast transcriptome (available at https://github.com/Jacobson-Lab/yeast_transcriptome_v5) using bowtie (Langmead et al. 2009), and transcript abundance was determined using RSEM (Li and Dewey 2011), as described previously (Mangkalaphiban et al. 2021). For intron-containing genes, spliced (“mRNA”) and unspliced (“pre-mRNA”) isoforms were indexed as separate entries.
Ribosome profiling libraries were preprocessed with adapter trimming and removal of reads aligned to non–protein-coding RNAs. Because some of the constructs were tagged at their amino termini, a fraction of reads recovered were from ribosomes associated with the Upf factor nascent peptide (Supplemental Table S1). Therefore, reads arising from UPF factor transcripts were discarded from these and all subsequent ribosome profiling libraries analyzed. PCR duplicates identified via the 4N barcodes on either side of the read were removed (Mangkalaphiban et al. 2021). Read processing statistics for ribosome profiling libraries are provided in Supplemental Table S1.
The remaining unique reads were further processed in two ways:
Transcript abundance was determined by RSEM either with all reads or a subset of reads of desired read lengths using a transcriptome containing pre-mRNA entries (rsem-calculate-expression –strandedness forward –fragment-length-mean [vary] –fragment-length-sd [vary] –seed-length 15 –bowtie-m 10). Reproducibility of biological replicate libraries is shown as a correlation matrix (Supplemental Fig. S11A) and PCA plot (Supplemental Fig. S11B).
Reads were aligned to a separate transcriptome where only spliced mRNAs were considered (bowtie -m 10 -n 2 -l 15). The resulting aligned reads were processed by R package riboWaltz (Lauria et al. 2018) for initial visual inspection and calculation of P-site offsets, which were manually checked and modified for accuracy (Supplemental Table S12) and used as the basis for subsequent reading frame calculations, periodicity, and other metagene plots. Analysis of ribosome footprints from both CHX treated and untreated cells and from total and Upf factor-associated ribosomes showed the footprints mapped primarily to coding regions; most P-sites mapped to the “0” reading frame in coding regions, and displayed 3-nt periodicity, indicative of translating ribosomes (Supplemental Fig. S7; Supplemental Fig. S10). As expected, CHX addition resulted in accumulation of ∼27–32 nt footprints over the start codon, as described previously (Ingolia 2010; Gerashchenko and Gladyshev 2014; Lareau et al. 2014). Replicates were averaged unless otherwise specified.
RNA-seq libraries were aligned to the transcriptome containing pre-mRNA entries and transcript abundance determined by RSEM (rsem-calculate-expression –strandedness reverse –fragment-length-mean 200 –fragment-length-sd 50 –bowtie-m 10) without any preprocessing steps. Reproducibility of biological replicate RNA-seq libraries is shown as a correlation matrix (Supplemental Fig. S12A) and PCA plot (Supplemental Fig. S12B).
Analyses of changes in transcript abundance
All analyses involving transcript abundance changes [changes in mRNA abundance: log2(FLAG-tagged/WT + EV); ribosome occupancy: log2(Total Ribo-Seq/RNA-seq); log2(IP/Total)] were performed in the R programming environment using R package DESeq2 (Love et al. 2014). The “expected_count” columns in the RSEM file output “isoforms.results” were used as input. Default parameters were used in all analyses with false discovery rate (FDR) to adjust P-value for multiple testing correction. For differential expression analysis between RNA-seq libraries, adjusted P-value cutoff of 0.01 was used to determine significant changes. For all other analyses, adjusted P-value cutoff of 0.05 was used to determine significant changes.
Analysis of A-site codon occupancy
Calculations of relative A-site codon occupancy values were adapted from Hussmann et al. (2015). For each mapped read, its P-site coordinate was determined by the offset provided in Supplemental Table S12. P-site coordinates were added by 3 to obtain A-site coordinates of the reads. For each mRNA sequence in the transcriptome that had any reads mapped to them, the coordinates of a codon of interest were determined. Reads that had their A-sites mapped within 60-codon window (−30 to +30 codon positions; −90 to +90 nt positions) of the codon were tabulated. After tabulating reads for each nucleotide position around the codon, reads were summed and divided by number of codon positions to obtain average read density. An entry with average density <0.1 (i.e., less than 1 read per 10 codons) was discarded from further analyses.
For codon-resolution analysis, reads at nucleotide positions that belong to the same codon position were summed and nucleotide positions were converted to codon positions. Relative occupancy at each position was calculated by dividing the number of reads at that position by the average read density. Relative occupancy of 1 would mean the A-site occupancy at that position is not different from the average across the 60-codon window, where relative occupancy higher and lower than 1 would mean enrichment and depletion, respectively, relative to the average. Then, mean relative occupancy for each codon position was calculated by averaging relative occupancy values of all occurrences of a codon in the transcriptome.
For comparison of mean relative occupancy between IP and Total ribosome samples, mean relative occupancy at each position from IP was divided by that of Total to get a ratio, and log2 transformed.
Codon optimality
We used the tRNA adaptation index (tAI), derived from tRNA gene copy numbers and wobble base-pairing penalty (dos Reis et al. 2004; Tuller et al. 2010b), as a measurement of codon optimality for a given codon identity. The codon optimality score for a coding sequence was calculated by taking the geometric mean of tAIs associated with all codons in the coding sequence (dos Reis et al. 2004).
Statistical analyses and data visualization
Due to the non-normal nature of the data, a two-tailed Wilcoxon's rank sum test with false discovery rate (FDR) for multiple testing correction was used to compare ribosome occupancy, coding sequence length, and codon optimality between IP-enriched, IP-depleted, and unchanged groups of mRNAs.
Fisher's exact test was performed by fisher.test() function and logistic regression by glm() function with “family” parameter = binomial(link = “logit”) in the R stats package.
R packages used for data preparation, statistical analyses, and visualization were DESeq2, riboWaltz, limma, stats, rstatix, MASS, ggplot2, ggpubr, ggrepel, ggh4x, scales, patchwork, Cairo, readxl, data.table, dplyr, and reshape2.
DATA DEPOSITION
All custom scripts have been made available at https://github.com/Jacobson-Lab/Upf1-ribosomes. Sequencing data that support this study have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) with the accession code GSE186795 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE186795). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (Perez-Riverol et al. 2019) with the data set identifier PXD029577 and 10.6019/PXD029577.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
COMPETING INTEREST STATEMENT
A.J. is cofounder, director, and Scientific Advisory Board chair of PTC Therapeutics Inc. All other authors declare no competing interests.
ACKNOWLEDGMENTS
This work was supported by a grant to A.J. (1R35GM122468) from the U.S. National Institutes of Health. We thank EiEi Min for providing ribosomes used early in the development of the selective ribosome profiling protocol; Alper Celik for his generous gifts of yeast strains and plasmids; Chan Wu, Alper Celik, EiEi Min, Scott Shaffer, Andrei Korostelev, Zhiping Weng, and Sean Ryder for helpful discussions; members of the UMass Chan Medical School Electron Microscopy Core Facility for electron micrographs; members of the UMass Chan Medical School Mass Spectrometry Core Facility for proteomics analyses; and the UMass Chan Medical School Molecular Biology Core Labs (RRID: SCR_018263) for RNA and sequencing library quality control analyses.
Author contributions: R.G. and A.J. conceived and designed the experiments, R.G. and K.M. carried out the experiments, R.B. and K.M. wrote data processing scripts, K.M., R.B., R.G., F.H., and A.J. analyzed the data, R.G., K.M., and A.J. wrote the paper with input from all authors, and A.J. obtained funding for the study.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079416.122.
Freely available online through the RNA Open Access option.
MEET THE FIRST AUTHORS
Robin Ganesan.

Kotchaphorn Mangkalaphiban.

Meet the First Author(s) is a new editorial feature within RNA, in which the first author(s) of research-based papers in each issue have the opportunity to introduce themselves and their work to readers of RNA and the RNA research community. Robin Ganesan and Kotchaphorn Mangkalaphiban are co-first authors of this paper, “Ribosome-bound Upf1 forms distinct 80S complexes and conducts mRNA surveillance.” Robin and Kotchaphorn both work in the lab of Allan Jacobson at UMass Chan Medical School. Robin is a Research Associate II, focusing on elucidating the mechanism by which the nonsense-mediated mRNA decay factors sense premature translation termination and trigger mRNA decay. Kotchaphorn is a graduate student, studying aspects of translation termination and their connections to nonsense-mediated mRNA decay, using ribosome profiling and other high-throughput sequencing methods.
What are the major results described in your paper and how do they impact this branch of the field?
Nonsense-mediated mRNA decay (NMD) is a process by which an encounter between a ribosome and a premature stop codon leads to rapid mRNA decay, and this process requires the functions of the NMD factors Upf1, Upf2, and Upf3. These factors comigrate with polyribosomes and also play a role in translation termination efficiency. While the biochemical properties of the core NMD factor Upf1 have been extensively studied, determining its function while bound to ribosomes has proven elusive. Here, we determined that Upf1 binds stochastically to translationally elongating ribosomes, that ribosome-bound Upf1 can form unique complexes that precede commitment to NMD, and that ATPase deficient Upf1 can interfere with normal termination events.
What led you to study RNA or this aspect of RNA science?
RG: I joined Allan's lab when the study of nonsense-mediated mRNA decay and the RNA field in general were still very young. RNA metabolism was particularly intriguing to me because little was known about it at the time, and the field was ripe for discovery. I have not been disappointed and am grateful to have seen this field blossom as it has over the last 30 years.
KM: My knowledge of translation termination was limited to the fact that three of the 64 codons are stop codons, signifying the end of translation. So, it was fascinating to learn about an mRNA decay pathway that takes care of mRNAs undergoing premature termination and to wonder how premature and normal termination are distinguished from each other. Coupled with my interest in working with sequencing data, I joined Allan's lab and Robin on this project to study translation-related aspects of this pathway at a transcriptome-wide level.
During the course of these experiments, were there any surprising results or particular difficulties that altered your thinking and subsequent focus?
RG: Because we were working with small amounts of input mRNA fragments for Ribo-seq libraries made from immunopurified ribosomes, we opted to use a protocol that eliminated gel size selection of RNA and cDNA. Fortuitously, this allowed us to recover a wide size range of ribosome protected fragments and led to our surprising discovery of the large ∼37–43 nt class of fragments. As a result, we followed up our original experiments with investigation of ATPase-deficient Upf1 as well as a WT Upf1 in a upf2Δ background and discovered that this fragment size class was dependent on Upf1 function but not on a functional NMD pathway.
What are some of the landmark moments that provoked your interest in science or your development as a scientist?
RG: I have to give original credit to my high school biology teacher who encouraged my interest in molecular biology in the early days of what was then called “genetic engineering.” During college, many faculty generously allowed me to work in their labs on independent research projects, but in particular Dr. Craig Giroux, who mentored and supported me during a time when there were few women in science. A particularly memorable moment in my career came when I was sequencing the ends of Drosophila chromosomes and saw for the first time a partial open reading frame extending to the very end of the telomere. This turned out to be the TART retrotransposon which functions in place of telomerase in this organism. The thrill of being the first person to see a novel process like that is unforgettable.
Are there specific individuals or groups who have influenced your philosophy or approach to science?
KM: Before graduate school, I had viewed science as something factual, almost unchanging. I loved science and math classes because they are methodical and there exist the “right” answers. Once I got the answers, I was done with homework. Of course, science is the discovery of the unknown, but it didn't hit me just how disconcerting the unknown is until graduate school. Suddenly, I went from knowing some things (answers to exam questions) to not knowing anything (in my research project). I would like to give credit to my PhD program as a whole for telling us very early and explicitly that we can, and should, challenge the current knowledge, and that for all of us, including experts in the field, there are many more things we don't know than what we do know. This should be exciting rather than scary. With this mindset change, I approach science as a lifelong learning process and am more willing to learn new things to get closer to an answer.
What are your subsequent near- or long-term career plans?
KM: I will be graduating soon, so I plan to continue my journey as a scientist in industry. I like analyzing sequencing data and finding patterns in big data. I hope to apply my growing interest and skills in this area to the development of drugs, assays, or diagnostics that would further the understanding of human health and the environment.
What were the strongest aspects of your collaboration as co-first authors?
RG: For me, the strongest aspect was the skill and insight of my collaborator Kotcha. Working with someone who not only has very strong bench skills but who is also an extremely meticulous and capable bioinformatician has been an invaluable asset. Our strengths have complemented each other synergistically, and I am very grateful to have had the opportunity to work with her.
KM: This is my first collaborative project, and I learned a lot from working with Robin. I may have offered a specific skill set, but without her insights into the biology and her ways of asking the right questions, we would not have been able to tease apart the data as much as we did together. It was a pleasure to have worked with her.
REFERENCES
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A. 2004. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432: 112–118. 10.1038/nature03060 [DOI] [PubMed] [Google Scholar]
- Atkin AL, Altamura N, Leeds P, Culbertson MR. 1995. The majority of yeast UPF1 co-localizes with polyribosomes in the cytoplasm. Mol Biol Cell 6: 611–625. 10.1091/mbc.6.5.611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkin AL, Schenkman LR, Eastham M, Dahlseid JN, Lelivelt MJ, Culbertson MR. 1997. Relationship between yeast polyribosomes and Upf proteins required for nonsense mRNA decay. J Biol Chem 272: 22163–22172. 10.1074/jbc.272.35.22163 [DOI] [PubMed] [Google Scholar]
- Becker AH, Oh E, Weissman JS, Kramer G, Bukau B. 2013. Selective ribosome profiling as a tool for studying the interaction of chaperones and targeting factors with nascent polypeptide chains and ribosomes. Nat Protoc 8: 2212–2239. 10.1038/nprot.2013.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm V, Kueckelmann S, Gerbracht JV, Kallabis S, Britto-Borges T, Altmuller J, Kruger M, Dieterich C, Gehring NH. 2021. SMG5-SMG7 authorize nonsense-mediated mRNA decay by enabling SMG6 endonucleolytic activity. Nat Commun 12: 3965. 10.1038/s41467-021-24046-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celik A. 2017. “mRNA decay pathways use translation fidelity and competing decapping complexes for substrate selection.” PhD thesis, Morningside Graduate School of Biomedical Sciences, UMass Chan Medical School. https://escholarship.umassmed.edu/cgi/viewcontent.cgi?article=1908&context=gsbs_diss. [Google Scholar]
- Celik A, Baker R, He F, Jacobson A. 2017. High-resolution profiling of NMD targets in yeast reveals translational fidelity as a basis for substrate selection. RNA 23: 735–748. 10.1261/rna.060541.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S, Jayachandran U, Bonneau F, Fiorini F, Basquin C, Domcke S, Le Hir H, Conti E. 2011. Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Mol Cell 41: 693–703. 10.1016/j.molcel.2011.02.010 [DOI] [PubMed] [Google Scholar]
- Colombo M, Karousis ED, Bourquin J, Bruggmann R, Muhlemann O. 2017. Transcriptome-wide identification of NMD-targeted human mRNAs reveals extensive redundancy between SMG6- and SMG7-mediated degradation pathways. RNA 23: 189–201. 10.1261/rna.059055.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K, Weng Y, Hagan KW, Peltz SW. 1995. Purification and characterization of the Upf1 protein: a factor involved in translation and mRNA degradation. RNA 1: 610–623. [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K, Ruiz-Echevarria MJ, Paushkin SV, Han X, Weng Y, Perlick HA, Dietz HC, Ter-Avanesyan MD, Peltz SW. 1998. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev 12: 1665–1677. 10.1101/gad.12.11.1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana A, Tuller T. 2014. The effect of tRNA levels on decoding times of mRNA codons. Nucleic Acids Res 42: 9171–9181. 10.1093/nar/gku646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehecq M, Decourty L, Namane A, Proux C, Kanaan J, Le Hir H, Jacquier A, Saveanu C. 2018. Nonsense-mediated mRNA decay involves two distinct Upf1-bound complexes. EMBO J 37: e99278. 10.15252/embj.201899278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doring K, Ahmed N, Riemer T, Suresh HG, Vainshtein Y, Habich M, Riemer J, Mayer MP, O'Brien EP, Kramer G, et al. 2017. Profiling Ssb-nascent chain interactions reveals principles of Hsp70-assisted folding. Cell 170: 298–311.e220. 10.1016/j.cell.2017.06.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Reis M, Savva R, Wernisch L. 2004. Solving the riddle of codon usage preferences: a test for translational selection. Nucleic Acids Res 32: 5036–5044. 10.1093/nar/gkh834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH. 2009. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol 16: 49–55. 10.1038/nsmb.1530 [DOI] [PubMed] [Google Scholar]
- Franks TM, Singh G, Lykke-Andersen J. 2010. Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense-mediated mRNA decay. Cell 143: 938–950. 10.1016/j.cell.2010.11.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan R, Leszyk J, Jacobson A. 2019. Selective profiling of ribosomes associated with yeast Upf proteins. Methods 155: 58–67. 10.1016/j.ymeth.2018.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerashchenko MV, Gladyshev VN. 2014. Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic Acids Res 42: e134. 10.1093/nar/gku671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Ganesan R, Amrani N, Jacobson A. 2010. Translational competence of ribosomes released from a premature termination codon is modulated by NMD factors. RNA 16: 1832–1847. 10.1261/rna.1987710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. 1988. New yeast–Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74: 527–534. 10.1016/0378-1119(88)90185-0 [DOI] [PubMed] [Google Scholar]
- Guydosh NR, Green R. 2017. Translation of poly(A) tails leads to precise mRNA cleavage. RNA 23: 749–761. 10.1261/rna.060418.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson CL, Videler H, Santos C, Ballesta JP, Robinson CV. 2004. Mass spectrometry of ribosomes from Saccharomyces cerevisiae: implications for assembly of the stalk complex. J Biol Chem 279: 42750–42757. 10.1074/jbc.M405718200 [DOI] [PubMed] [Google Scholar]
- He F, Jacobson A. 1995. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev 9: 437–454. 10.1101/gad.9.4.437 [DOI] [PubMed] [Google Scholar]
- He F, Jacobson A. 2001. Upf1p, Nmd2p, and Upf3p regulate the decapping and exonucleolytic degradation of both nonsense-containing mRNAs and wild-type mRNAs. Mol Cell Biol 21: 1515–1530. 10.1128/MCB.21.5.1515-1530.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Jacobson A. 2015. Nonsense-mediated mRNA decay: degradation of defective transcripts is only part of the story. Annu Rev Genet 49: 339–366. 10.1146/annurev-genet-112414-054639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Peltz SW, Donahue JL, Rosbash M, Jacobson A. 1993. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1-mutant. Proc Natl Acad Sci 90: 7034–7038. 10.1073/pnas.90.15.7034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Brown AH, Jacobson A. 1997. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol Cell Biol 17: 1580–1594. 10.1128/MCB.17.3.1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Ganesan R, Jacobson A. 2013. Intra- and intermolecular regulatory interactions in Upf1, the RNA helicase central to nonsense-mediated mRNA decay in yeast. Mol Cell Biol 33: 4672–4684. 10.1128/MCB.01136-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Celik A, Wu C, Jacobson A. 2018. General decapping activators target different subsets of inefficiently translated mRNAs. Elife 7: e34409. 10.7554/eLife.34409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Wu C, Jacobson A. 2022. Dcp2 C-terminal cis-binding elements control selective targeting of the decapping enzyme by forming distinct decapping complexes. Elife 11: 74410. 10.7554/eLife.74410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrick D, Parker R, Jacobson A. 1990. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol Cell Biol 10: 2269–2284. 10.1128/mcb.10.5.2269-2284.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg JR, Goff SP. 2010. Upf1 senses 3′UTR length to potentiate mRNA decay. Cell 143: 379–389. 10.1016/j.cell.2010.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Kashima I, Fauser M, Sauliere J, Izaurralde E. 2008. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA 14: 2609–2617. 10.1261/rna.1386208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt JA, Robertson AD, Burge CB. 2013. Global analyses of UPF1 binding and function reveal expanded scope of nonsense-mediated mRNA decay. Genome Res 23: 1636–1650. 10.1101/gr.157354.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussmann JA, Patchett S, Johnson A, Sawyer S, Press WH. 2015. Understanding biases in ribosome profiling experiments reveals signatures of translation dynamics in yeast. PLoS Genet 11: e1005732. 10.1371/journal.pgen.1005732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT. 2010. Genome-wide translational profiling by ribosome footprinting. Methods Enzymol 470: 119–142. 10.1016/S0076-6879(10)70006-9 [DOI] [PubMed] [Google Scholar]
- Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE. 2008. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J 27: 736–747. 10.1038/emboj.2008.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammers K, Cole RN, Tiengwe C, Ruczinski I. 2015. Detecting significant changes in protein abundance. EuPA Open Proteom 7: 11–19. 10.1016/j.euprot.2015.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S. 2006. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev 20: 355–367. 10.1101/gad.1389006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74: 5383–5392. 10.1021/ac025747h [DOI] [PubMed] [Google Scholar]
- Kim YK, Maquat LE. 2019. UPFront and center in RNA decay: UPF1 in nonsense-mediated mRNA decay and beyond. RNA 25: 407–422. 10.1261/rna.070136.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki T, Maquat LE. 2013. Rules that govern UPF1 binding to mRNA 3′ UTRs. Proc Natl Acad Sci 110: 3357–3362. 10.1073/pnas.1219908110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki T, Miyoshi K, Myers JR, Maquat LE. 2018. NMD-degradome sequencing reveals ribosome-bound intermediates with 3′-end non-templated nucleotides. Nat Struct Mol Biol 25: 940–950. 10.1038/s41594-018-0132-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki T, Popp MW, Maquat LE. 2019. Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat Rev Mol Cell Biol 20: 406–420. 10.1038/s41580-019-0126-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau LF, Hite DH, Hogan GJ, Brown PO. 2014. Distinct stages of the translation elongation cycle revealed by sequencing ribosome-protected mRNA fragments. Elife 3: e01257. 10.7554/eLife.01257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauria F, Tebaldi T, Bernabo P, Groen EJN, Gillingwater TH, Viero G. 2018. riboWaltz: optimization of ribosome P-site positioning in ribosome profiling data. PLoS Comput Biol 14: e1006169. 10.1371/journal.pcbi.1006169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeds P, Peltz SW, Jacobson A, Culbertson MR. 1991. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev 5: 2303–2314. 10.1101/gad.5.12a.2303 [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323. 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh B, Jonas S, Izaurralde E. 2013. The SMG5-SMG7 heterodimer directly recruits the CCR4-NOT deadenylase complex to mRNAs containing nonsense codons via interaction with POP2. Genes Dev 27: 2125–2138. 10.1101/gad.226951.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Perrote A, Castano R, Melero R, Zamarro T, Kurosawa H, Ohnishi T, Uchiyama A, Aoyagi K, Buchwald G, Kataoka N, et al. 2016. Human nonsense-mediated mRNA decay factor UPF2 interacts directly with eRF3 and the SURF complex. Nucleic Acids Res 44: 1909–1923. 10.1093/nar/gkv1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losson R, Lacroute F. 1979. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc Natl Acad Sci 76: 5134–5137. 10.1073/pnas.76.10.5134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangkalaphiban K, He F, Ganesan R, Wu C, Baker R, Jacobson A. 2021. Transcriptome-wide investigation of stop codon readthrough in Saccharomyces cerevisiae. PLoS Genet 17: e1009538. 10.1371/journal.pgen.1009538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus DA, Jacobson A. 1999. Linking mRNA turnover and translation: assessing the polyribosomal association of mRNA decay factors and degradative intermediates. Methods 17: 28–37. 10.1006/meth.1998.0704 [DOI] [PubMed] [Google Scholar]
- Maquat LE. 2000. Nonsense-mediated mRNA decay in mammalian cells: a splicing-dependent means to down-regulate the levels of mRNAs that prematurely terminate translation. In Translational control of gene expression, 2nd ed (ed. Sonenberg N, et al.), pp. 849–868. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Min EE, Roy B, Amrani N, He F, Jacobson A. 2013. Yeast Upf1 CH domain interacts with Rps26 of the 40S ribosomal subunit. RNA 19: 1105–1115. 10.1261/rna.039396.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D, Parker R. 1994. Premature translational termination triggers mRNA decapping. Nature 370: 578–581. 10.1038/370578a0 [DOI] [PubMed] [Google Scholar]
- Nelson JO, Forster D, Frizzell KA, Luschnig S, Metzstein MM. 2018. Multiple nonsense-mediated mRNA processes require Smg5 in Drosophila. Genetics 209: 1073–1084. 10.1534/genetics.118.301140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75: 4646–4658. 10.1021/ac0341261 [DOI] [PubMed] [Google Scholar]
- Nott A, Le Hir H, Moore MJ. 2004. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev 18: 210–222. 10.1101/gad.1163204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottens F, Boehm V, Sibley CR, Ule J, Gehring NH. 2017. Transcript-specific characteristics determine the contribution of endo- and exonucleolytic decay pathways during the degradation of nonsense-mediated decay substrates. RNA 23: 1224–1236. 10.1261/rna.059659.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, et al. 2019. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47: D442–D450. 10.1093/nar/gky1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presnyak V, Alhusaini N, Chen YH, Martin S, Morris N, Kline N, Olson S, Weinberg D, Baker KE, Graveley BR, et al. 2015. Codon optimality is a major determinant of mRNA stability. Cell 160: 1111–1124. 10.1016/j.cell.2015.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan A, Chen YH, Martin S, Alhusaini N, Green R, Coller J. 2016. The DEAD-Box protein Dhh1p couples mRNA decay and translation by monitoring codon optimality. Cell 167: 122–132.e129. 10.1016/j.cell.2016.08.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller AP, Zinshteyn B, Enam SU, Green R. 2018. Directed hydroxyl radical probing reveals Upf1 binding to the 80S ribosomal E site rRNA at the L1 stalk. Nucleic Acids Res 46: 2060–2073. 10.1093/nar/gkx1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. 2011. Global quantification of mammalian gene expression control. Nature 473: 337–342. 10.1038/nature10098 [DOI] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. 2013. Corrigendum: global quantification of mammalian gene expression control. Nature 495: 126–127. 10.1038/nature11848 [DOI] [PubMed] [Google Scholar]
- Serdar LD, Whiteside DL, Baker KE. 2016. ATP hydrolysis by UPF1 is required for efficient translation termination at premature stop codons. Nat Commun 7: 14021. 10.1038/ncomms14021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serdar LD, Whiteside DL, Nock SL, McGrath D, Baker KE. 2020. Inhibition of post-termination ribosome recycling at premature termination codons in UPF1 ATPase mutants. Elife 9: e57834. 10.7554/eLife.57834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Wu J, Nilges BS, Leidel SA. 2021. Humans and other commonly used model organisms are resistant to cycloheximide-mediated biases in ribosome profiling experiments. Nat Commun 12: 5094. 10.1038/s41467-021-25411-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. 10.1093/genetics/122.1.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3. 10.2202/1544-6115.1027 [DOI] [PubMed] [Google Scholar]
- Tuller T, Carmi A, Vestsigian K, Navon S, Dorfan Y, Zaborske J, Pan T, Dahan O, Furman I, Pilpel Y. 2010a. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell 141: 344–354. 10.1016/j.cell.2010.03.031 [DOI] [PubMed] [Google Scholar]
- Tuller T, Waldman YY, Kupiec M, Ruppin E. 2010b. Translation efficiency is determined by both codon bias and folding energy. Proc Natl Acad Sci 107: 3645–3650. 10.1073/pnas.0909910107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S, Herrmannova A, Hronova V, Gunisova S, Sen ND, Hannan RD, Hinnebusch AG, Shirokikh NE, Preiss T, Valasek LS. 2020. Selective translation complex profiling reveals staged initiation and co-translational assembly of initiation factor complexes. Mol Cell 79: 546–560.e547. 10.1016/j.molcel.2020.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Czaplinski K, Rao Y, Peltz SW. 2001. The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J 20: 880–890. 10.1093/emboj/20.4.880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CC, Zinshteyn B, Wehner KA, Green R. 2019. High-resolution ribosome profiling defines discrete ribosome elongation states and translational regulation during cellular stress. Mol Cell 73: 959–970.e955. 10.1016/j.molcel.2018.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Roy B, He F, Yan K, Jacobson A. 2020. Poly(A)-binding protein regulates the efficiency of translation termination. Cell Rep 33: 108399. 10.1016/j.celrep.2020.108399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa H, Larance M, Harney DJ, Sundaramoorthy R, Ly T, Owen-Hughes T, Lamond AI. 2018. Efficient analysis of mammalian polysomes in cells and tissues using Ribo Mega-SEC. Elife 7: e36530. 10.7554/eLife.36530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Welch EM, Hogan K, Brown AH, Peltz SW, Jacobson A. 1997. Polysome-associated mRNAs are substrates for the nonsense-mediated mRNA decay pathway in Saccharomyces cerevisiae. RNA 3: 234–244. [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Chen YM, Li Y, Wang J, Chen S, Gao N, Qian W. 2021. Disome-seq reveals widespread ribosome collisions that promote cotranslational protein folding. Genome Biol 22: 16. 10.1186/s13059-020-02256-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zund D, Gruber AR, Zavolan M, Muhlemann O. 2013. Translation-dependent displacement of UPF1 from coding sequences causes its enrichment in 3′ UTRs. Nat Struct Mol Biol 20: 936–943. 10.1038/nsmb.2635 [DOI] [PubMed] [Google Scholar]