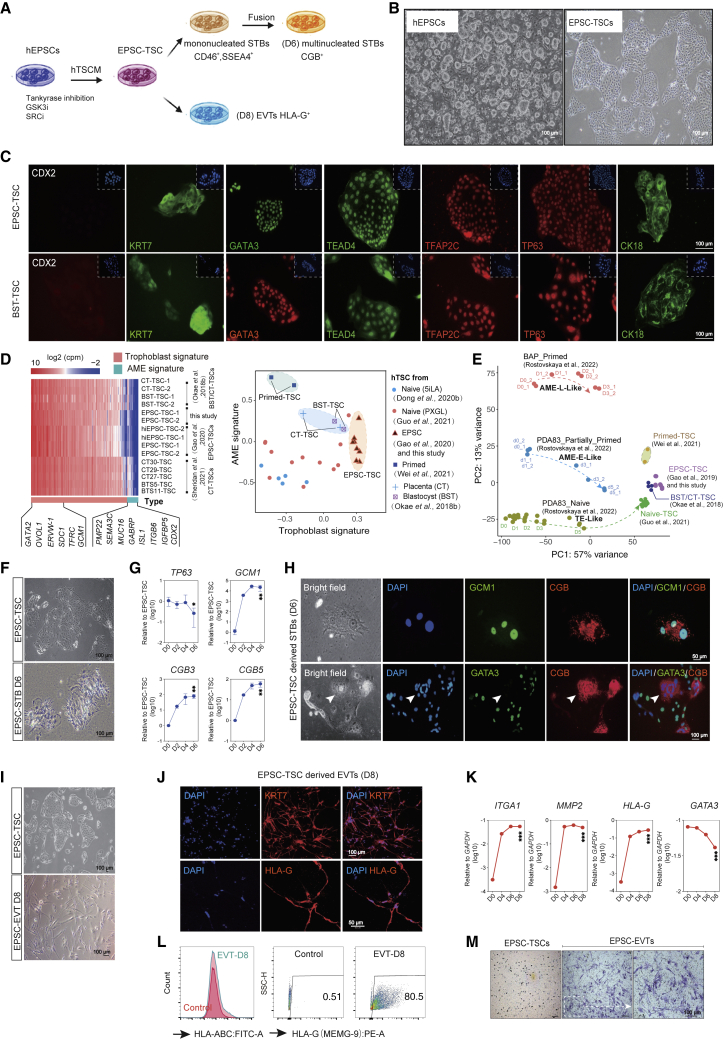
Figure 1.
Generation of human trophoblast stem cells (TSCs) and trophoblast subtypes from hEPSCs
(A) Schematic diagram of sequential generation of hTSCs, STBs, and EVTs from hEPSCs.
(B) Bright-field images of hEPSCs and EPSC-TSCs. Scale bars: 100 μm.
(C) Immunofluorescence-stained EPSC-TSCs and human blastocyst-derived hTSCs (BST-TSC).35 Scale bars, 100 μm.
(D) Left: RNA-seq analysis of human trophoblast and putative AME signature gene (Table S1) expression in EPSC- and BST/CT-TSC and five human placenta-derived CT-TSCs.36 Right: Scatterplot for gene set scores for human trophoblast and putative AME signature genes (Table S1) in hTSCs of various origins.
(E) PCA for comparison of hTSCs of in vivo (BST/CT) and in vitro origins (naive, primed, and EPSC). Datasets include in vitro differentiation of naive hPSCs to TSCs (D0–D5), primed hPSCs to AME-L (late)-like cells (D0–D3), and partially primed hPSCs to AME-E (early)-like (D0–D5).37 The dashed lines and arrows indicate the differentiation trajectories.
(F) Bright-field images of EPSC-TSCs differentiating toward STBs (STB-D6). Scale bars, 100 μm.
(G) Gene expression (qRT-PCR) during EPSC-TSC differentiation toward STBs. Data are mean ± SD; n = 3 biological replicates. Student’s t test, ∗p < 0.05, ∗∗p < 0.01.
(H) Immunostaining of STBs-D6 for GCM1 and CGB (top) and GATA3 (bottom). Arrows indicate the lack of GATA3 in a multinucleated CGB+ mature STB. Scale bars, 50 or 100 μm as indicated.
(I) Bright-field images of EPSC-TSCs differentiating toward EVT-D8. Scale bars, 100 μm.
(J) Immunostaining of EVT-D8 for KRT7 and HLA-G. Scale bars, 50 or 100 μm as indicated.
(K) Gene expression (qRT-PCR) during EPSC-TSC differentiation toward EVTs. Data are mean ± SD; n = 3 biological replicates. Student’s t test, ∗∗∗p < 0.001.
(L) Flow cytometric analysis of HLA-A, -B, -C and HLA-G on EVT-D8. EPSC-TSCs were used as the control.
(M) Invasiveness of EVT-D8 in the transwell assay. The invading/migrating cells on the lower surface were stained with crystal violet. Scale bar, 100 μm.