Figure 2.
Trophoblasts derived from hEPSCs resemble those in human peri-implantation embryos and placenta
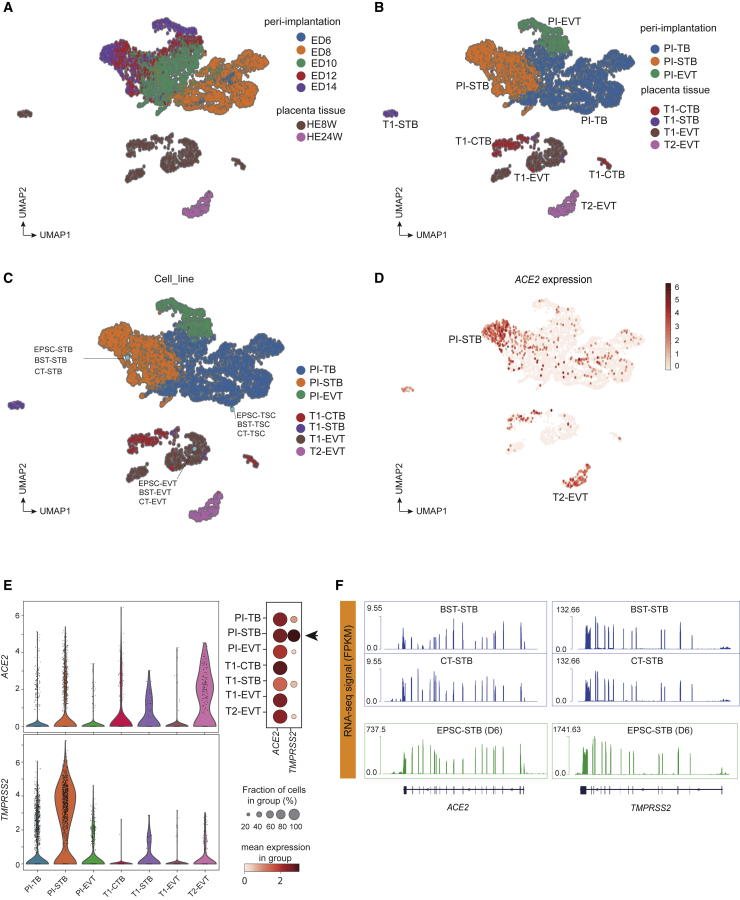
(A and B) UMAP analysis of scRNA-seq data of cells from in vitro-cultured peri-implantation (PI) embryo stages (top) and of cells from first (T1)- and second (T2)-trimester placenta (bottom). Cells are colored by developmental time points (ED6–14, embryonic day 6–14; HE8W/HE24W, placenta trophoblasts at 8 or 24 weeks of gestation, corresponding to first or second trimester) in (A) and trophoblast subtypes at each stage in (B).
(C) Mapping of in vitro human trophoblast cell bulk RNA-seq data to the peri-implantation and placental trophoblast clusters in (B). The in vitro cells are highlighted in light blue filled circles.
(D) Expression of ACE2 in peri-implantation and placenta trophoblast scRNA-seq clusters.
(E) Left: violin plots of ACE2 and TMPRSS2 expression (log-transformed transcripts per million [TPM]). Right: dot plot of ACE2 and TMPRSS2 expression levels in ACE2-positive cells, categorized according to stage and cell lineage. Dots are colored according to the mean expression value in each category and dot size indicates the percentage of ACE2- or TMPRSS2-positive cells from each category that expresses ACE2.
(F) RNA-seq signals of ACE2 and TMPRSS2 in BST/CT-STBs and EPSC-STBs. The ACE2 and TMPRSS2 genomic loci are plotted at the bottom, where each vertical bar represents an exon, and the transcription direction is from right to left.